Chapter 2 igraph package
2.1 Introduction
2.1.1 igraph vs statnet
](images/igrastatnet.png)
Figure 2.1: igraph versus statnet from Shizuka Lab
2.1.2 References
- Official website (handbook): http://igraph.org/r/
- Tutorial: http://kateto.net/networks-r-igraph
- Book: https://sites.fas.harvard.edu/~airoldi/pub/books/BookDraft-CsardiNepuszAiroldi2016.pdf
- Datasets:
2.1.3 Preparation
#install.packages("igraph")
#install.packages("igraphdata")
library(igraph)
library(igraphdata)2.2 Create networks and basics concepts
2.2.1 Outline
- Basic introduction on network analysis using R.
- R package
igraph- create networks (predifined structures; specific graphs; graph models; adjustments)
- Edge, vertex and network attributes
- Network and node descriptions
- R package
statnet(ERGM,…)
- R package
- Collecting network data
- Web API requesting (Twitter, Reddit, IMDB, or more)
- Useful websites (SNAP, or more)
- Visualization
- static networks and dynamic networks
- Network analysis
2.2.2 Create simple networks
graph(edges,n,directed,isolates)graph_from_literal
2.2.2.1 graph(edges,n,directed,isolates)
an undirected graph with 3 edges:
g1 <- graph( edges=c(1,2, 2,3, 3,1), n=3, directed=F )
plot(g1) 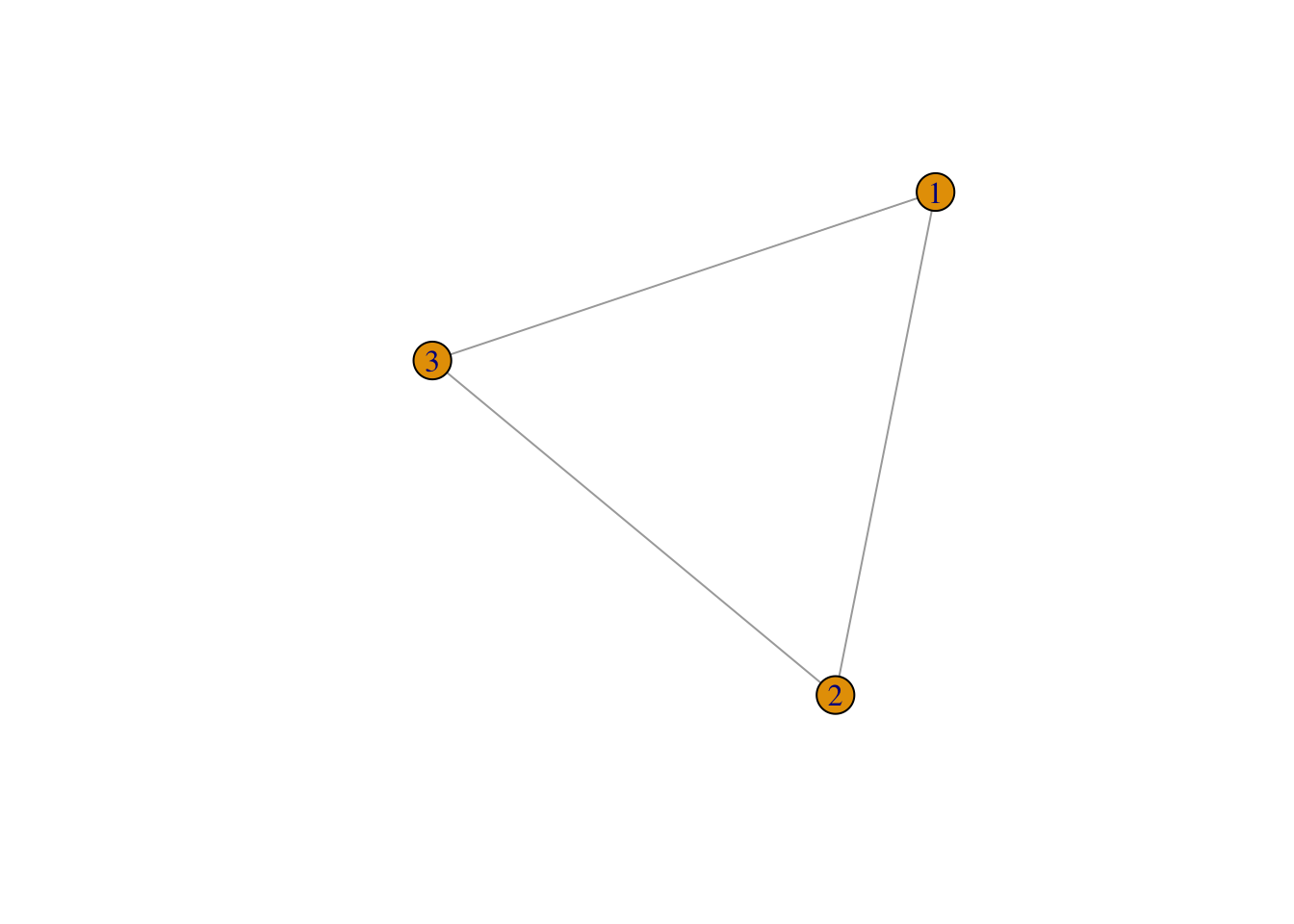
n can be greater than number of vertices in the edge list
g2 <- graph( edges=c(1,2, 2,3, 3,1), n=10 ) # now with 10 vertices, and directed by default
plot(g2) 
named vertices
g3 <- graph( c("John", "Jim", "Jim", "Jill", "Jill", "John"))
# When the edge list has vertex names, the number of nodes is not needed
plot(g3)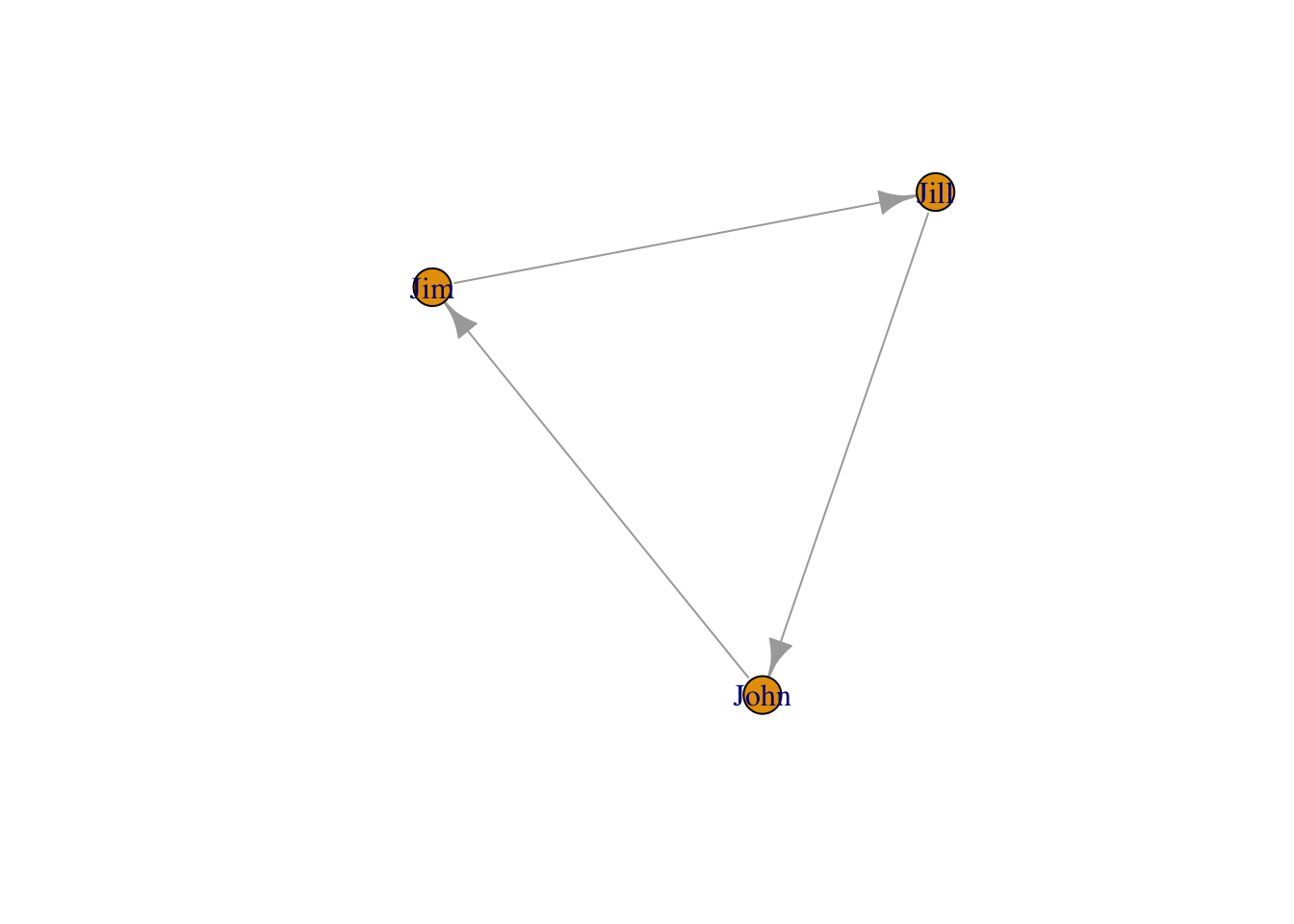
named vertices without edges
g4 <- graph( c("John", "Jim", "Jim", "Jack", "Jim", "Jack", "John", "John"),
isolates=c("Jesse", "Janis", "Jennifer", "Justin") )
# In named graphs we can specify isolates by providing a list of their names.
set.seed(1)
plot(g4, edge.arrow.size=.5, vertex.color="gold", vertex.size=15,
vertex.frame.color="gray", vertex.label.color="black",
vertex.label.cex=1.5, vertex.label.dist=2, edge.curved=0.2) 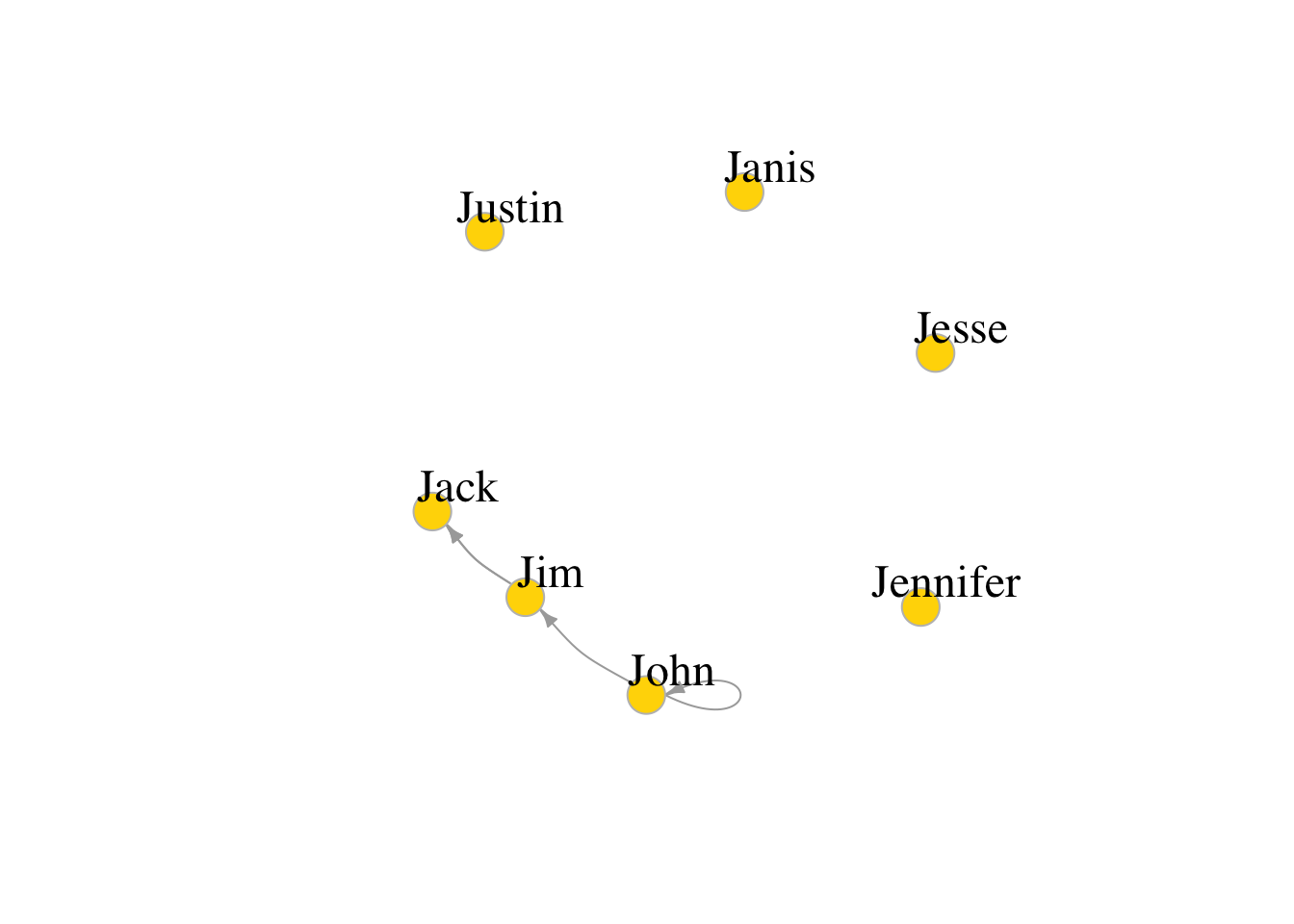
2.2.2.2 graph_from_literal
Small graphs can also be generated with a description of this kind:
- ‘-’ for undirected tie, “+-’ or”-+" for directed ties pointing left & right,
- “++” for a symmetric tie, and “:” for sets of vertices
plot(graph_from_literal(a---b, b---c)) # the number of dashes doesn't matter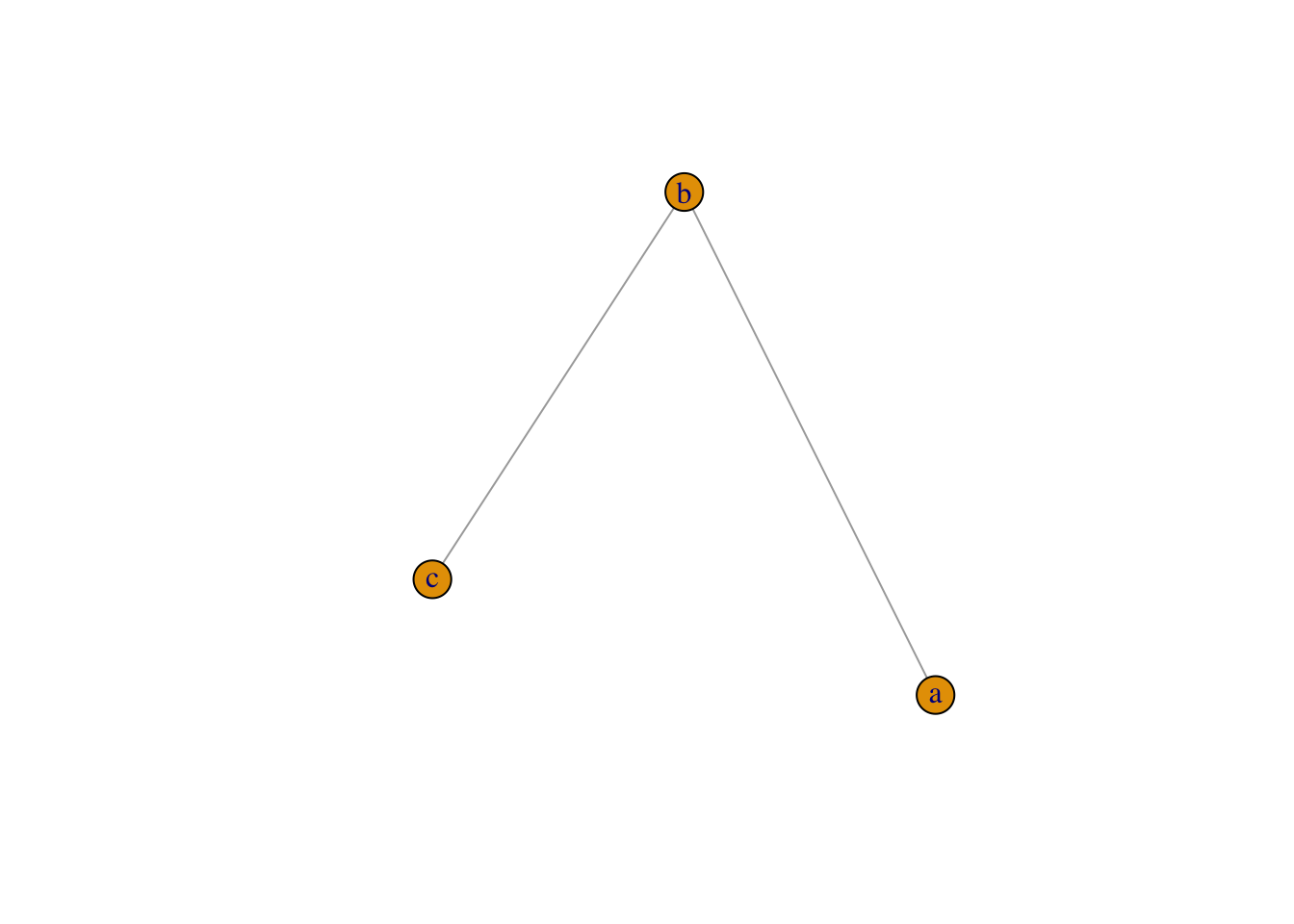
plot(graph_from_literal(a--+b, b+--c))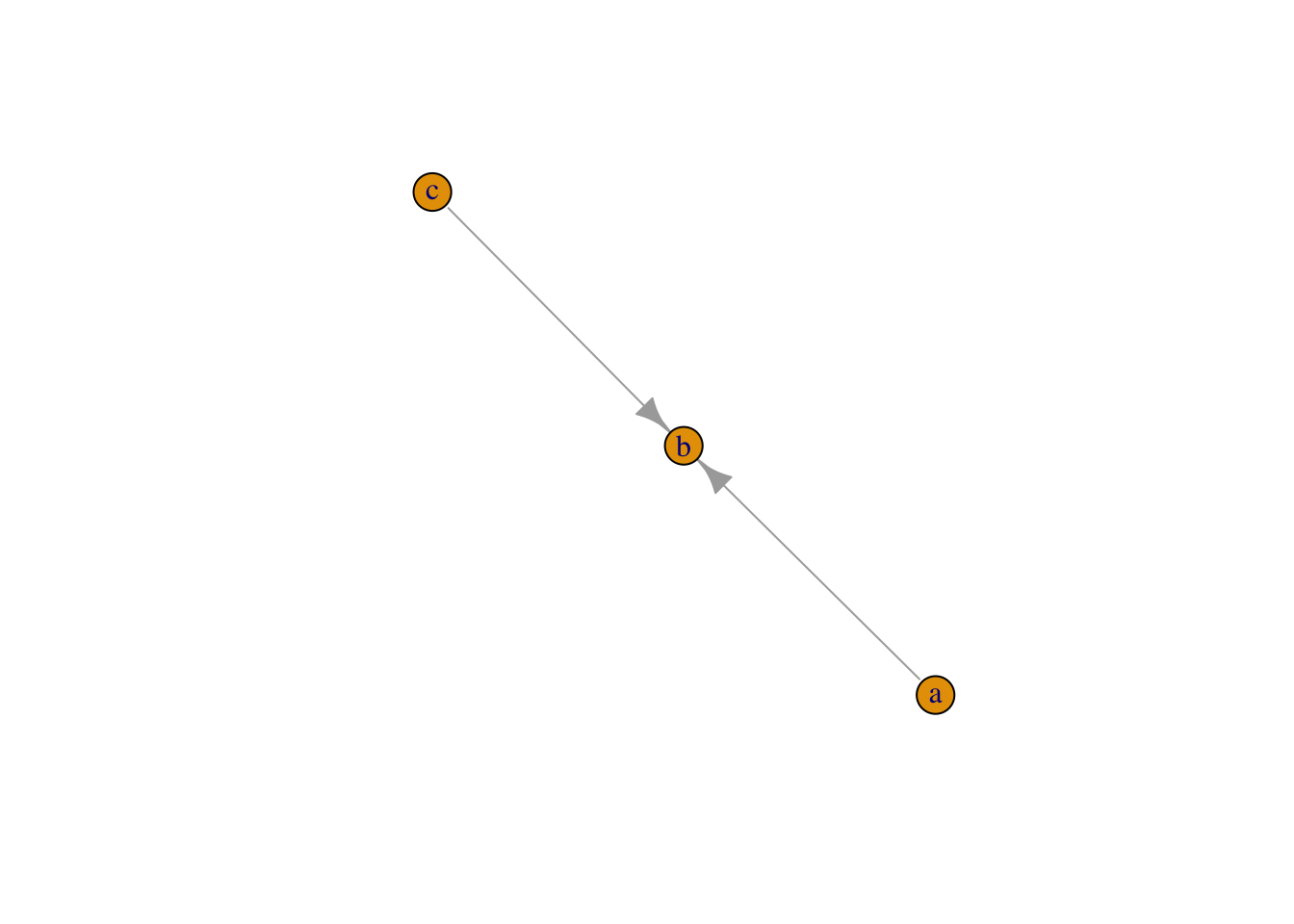
plot(graph_from_literal(a+-+b, b+-+c)) 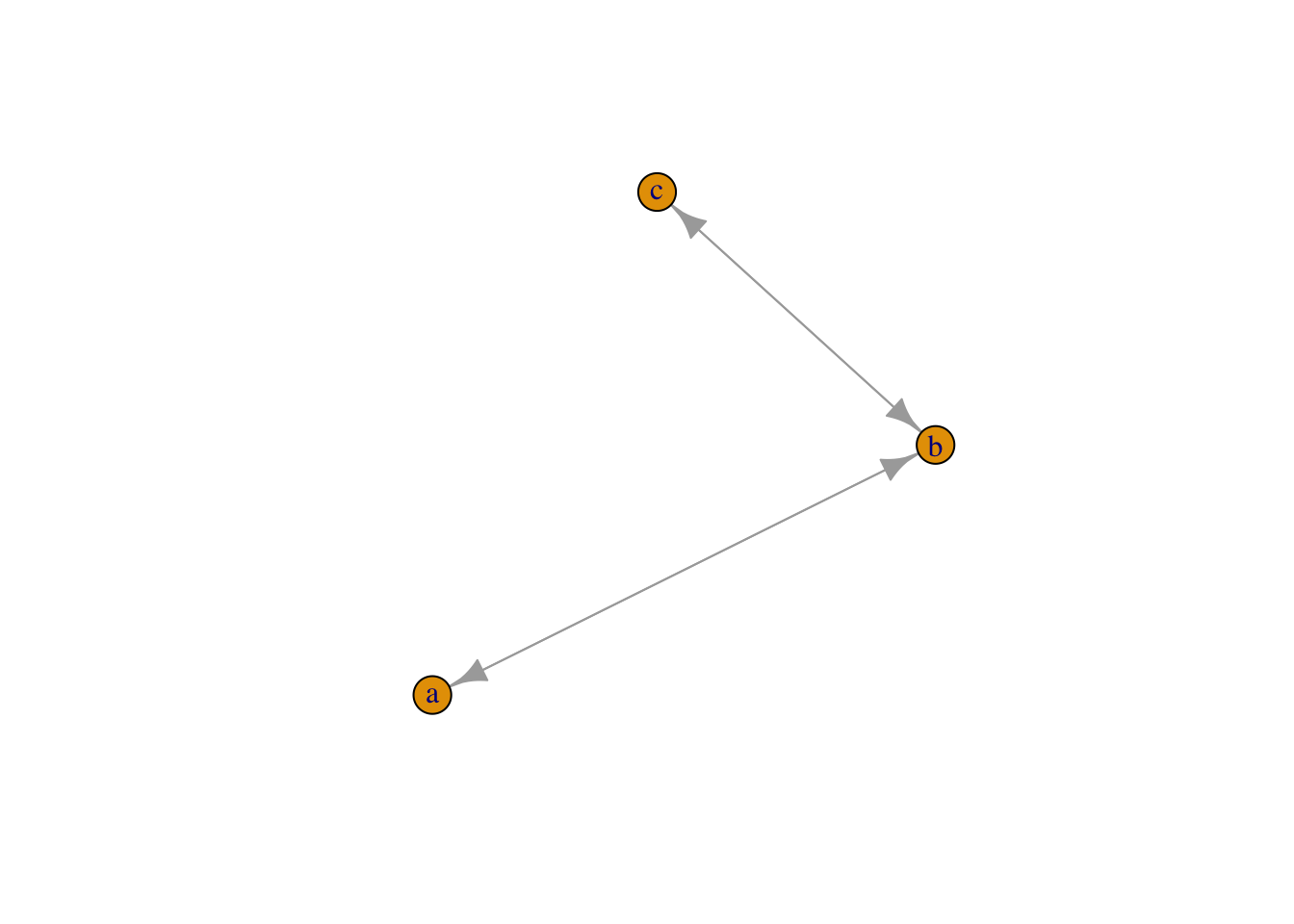
a:b:c using colon to connect abc as a whole group. Each vertex within group a:b:c is connected to each vertex within group c:d:e
plot(graph_from_literal(a:b:c---c:d:e))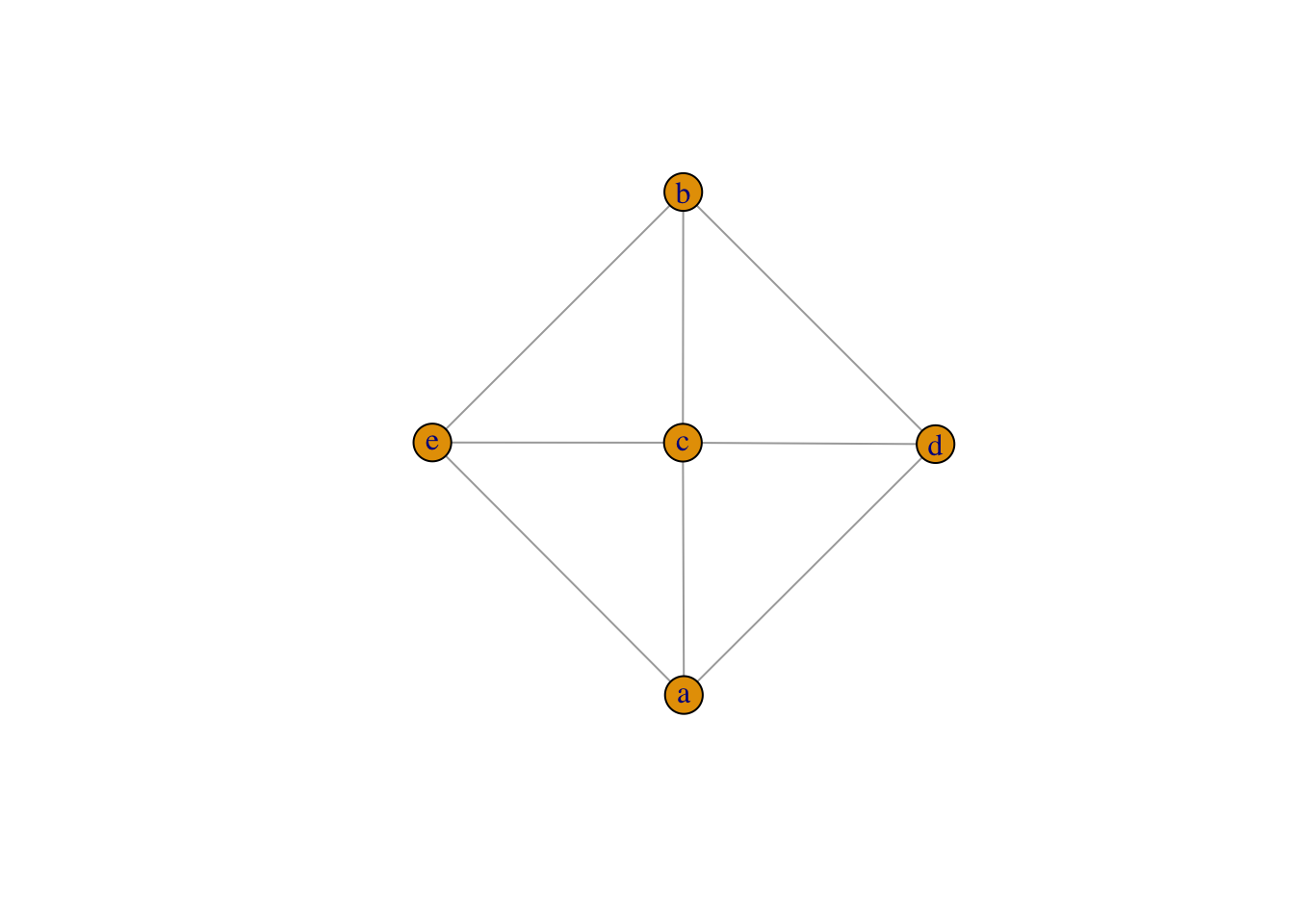
plot(graph_from_literal(a--b:c:d))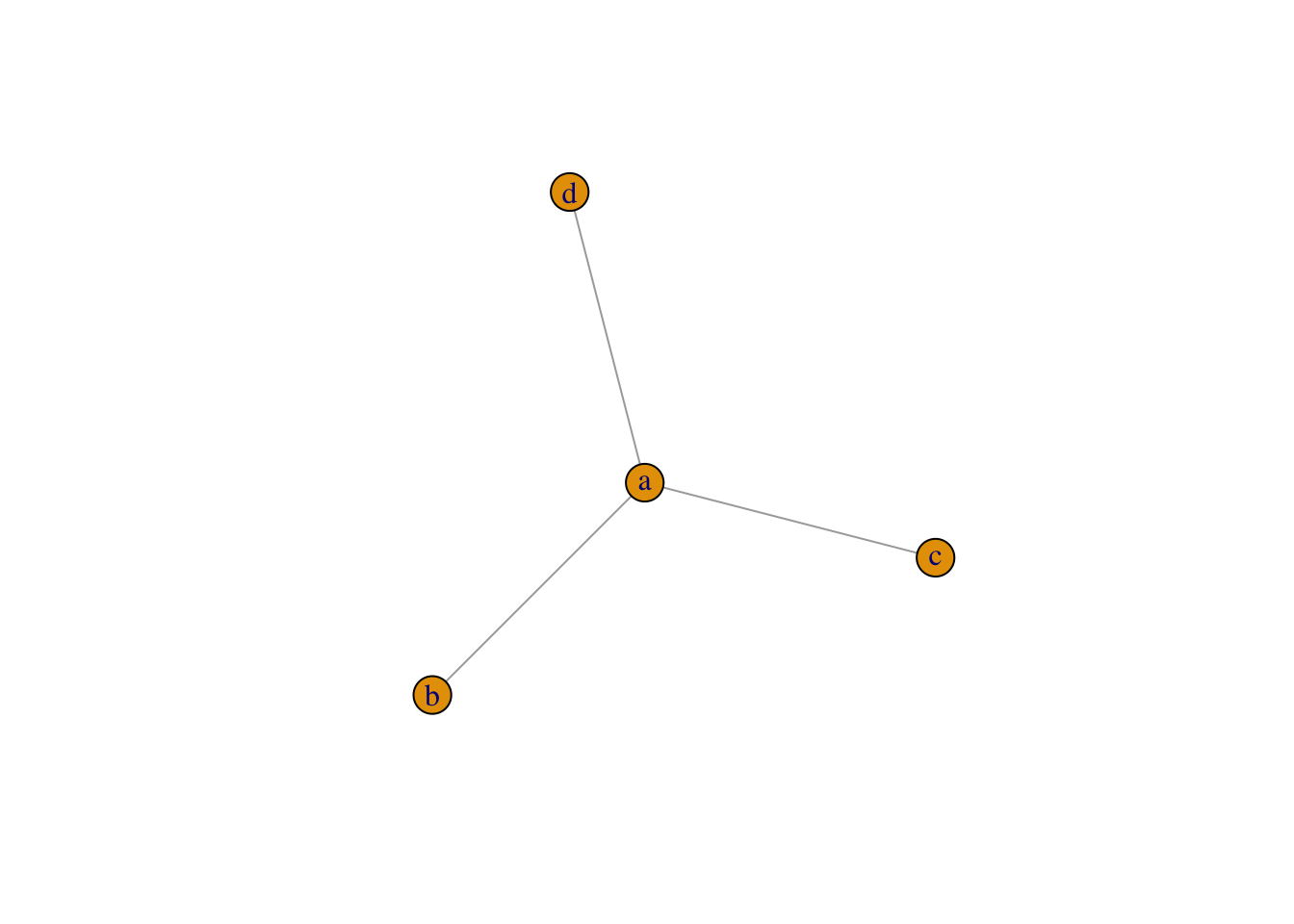
plot(graph_from_literal(a:e--b:c:d))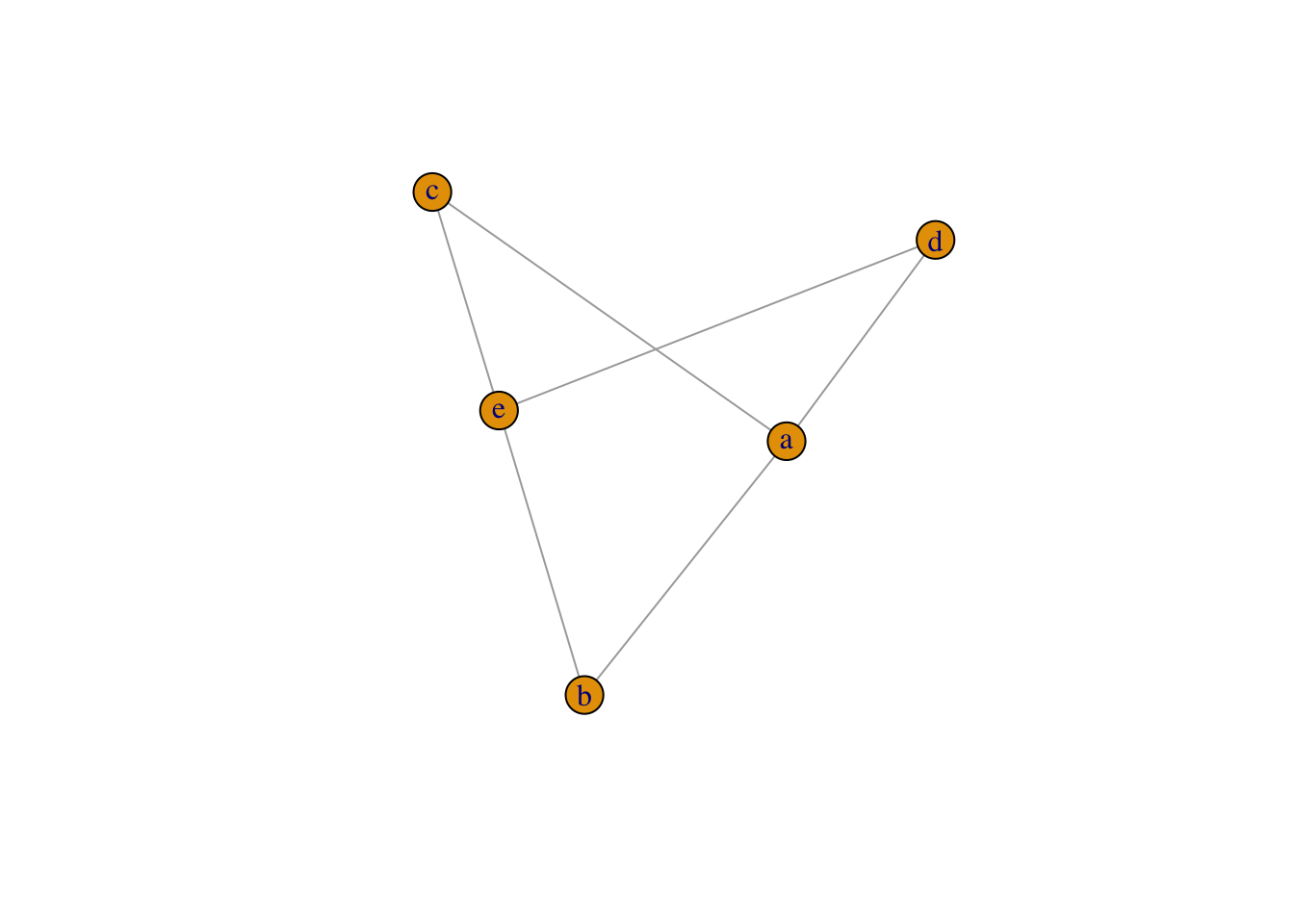
2.2.3 Creating specific graphs and graph models
- Specific graph
make_empty_graphmake_full_graphmake_treemake_starmake_ring
- Graph models
sample_gnmErdos-Renyi random graphsample_gnpErdos-Renyi with G(n,p) specificationsample_smallworldWatts-Strogatz small-world modelsample_paBarabasi-Albert preferential attachment model for scale-free graphs
2.2.3.1 Empty graph
eg <- make_empty_graph(40)
plot(eg, vertex.size=10, vertex.label=NA)
2.2.3.2 Full graph
fg <- make_full_graph(40)
plot(fg, vertex.size=10, vertex.label=NA)
2.2.3.3 Tree graph
tr <- make_tree(40, children = 3, mode = "undirected")
plot(tr, vertex.size=10, vertex.label=NA) 
2.2.3.4 Star graph
st <- make_star(40)
plot(st, vertex.size=10, vertex.label=NA) 
2.2.3.5 Ring graph
rn <- make_ring(40)
plot(rn, vertex.size=10, vertex.label=NA)
2.2.3.6 Erdos-Renyi random graph
‘n’ is number of nodes, ‘m’ is the number of edges
er <- sample_gnm(n=100, m=40) ####can also use erdos.renyi.game
## options include directed= and loops=
plot(er, vertex.size=6, vertex.label=NA) 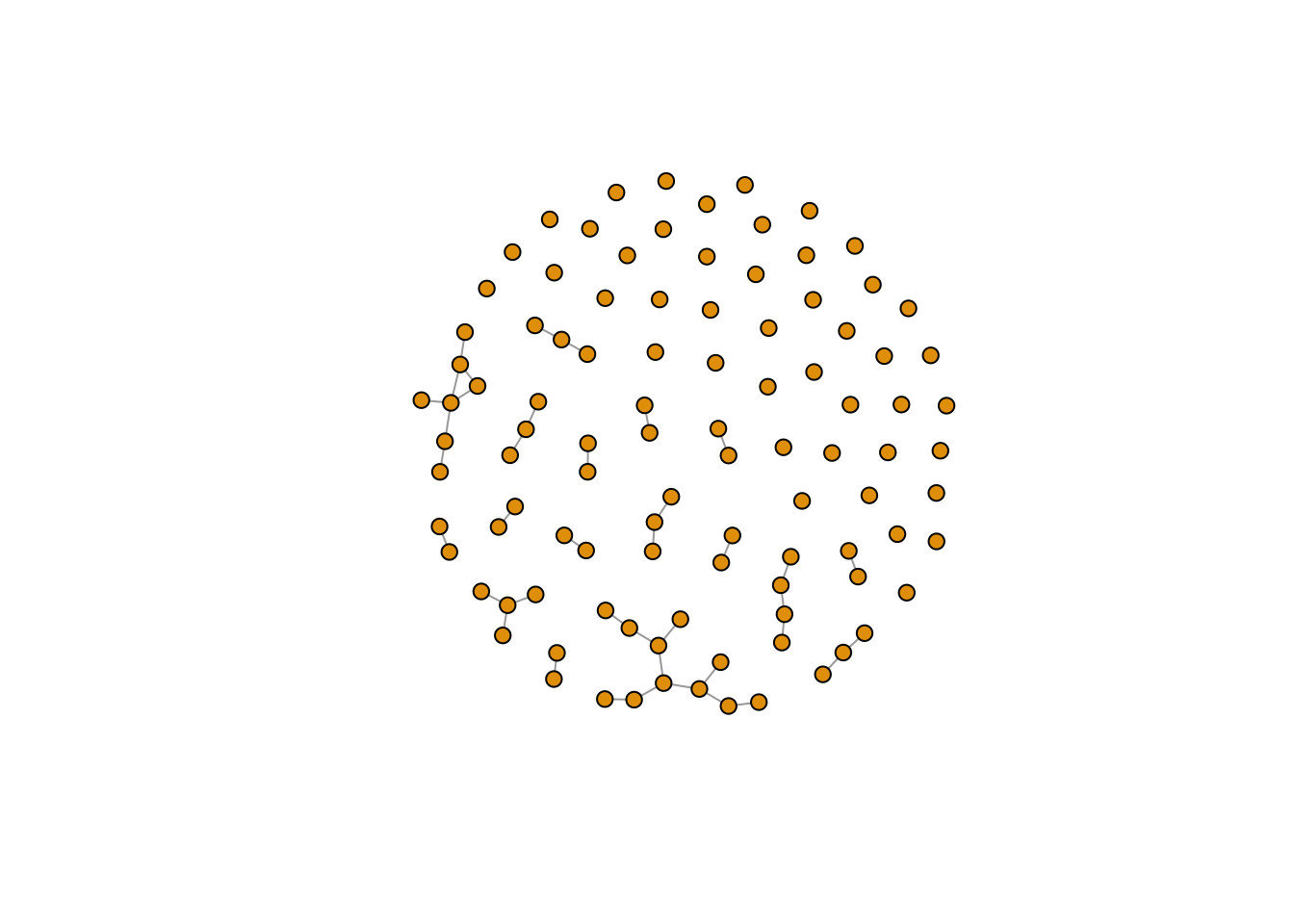
2.2.3.7 Erdos-Renyi with G(n,p) specification
er <- sample_gnp(n=100, p=.02) ####can also use erdos.renyi.game
plot(er, vertex.size=6, vertex.label=NA) 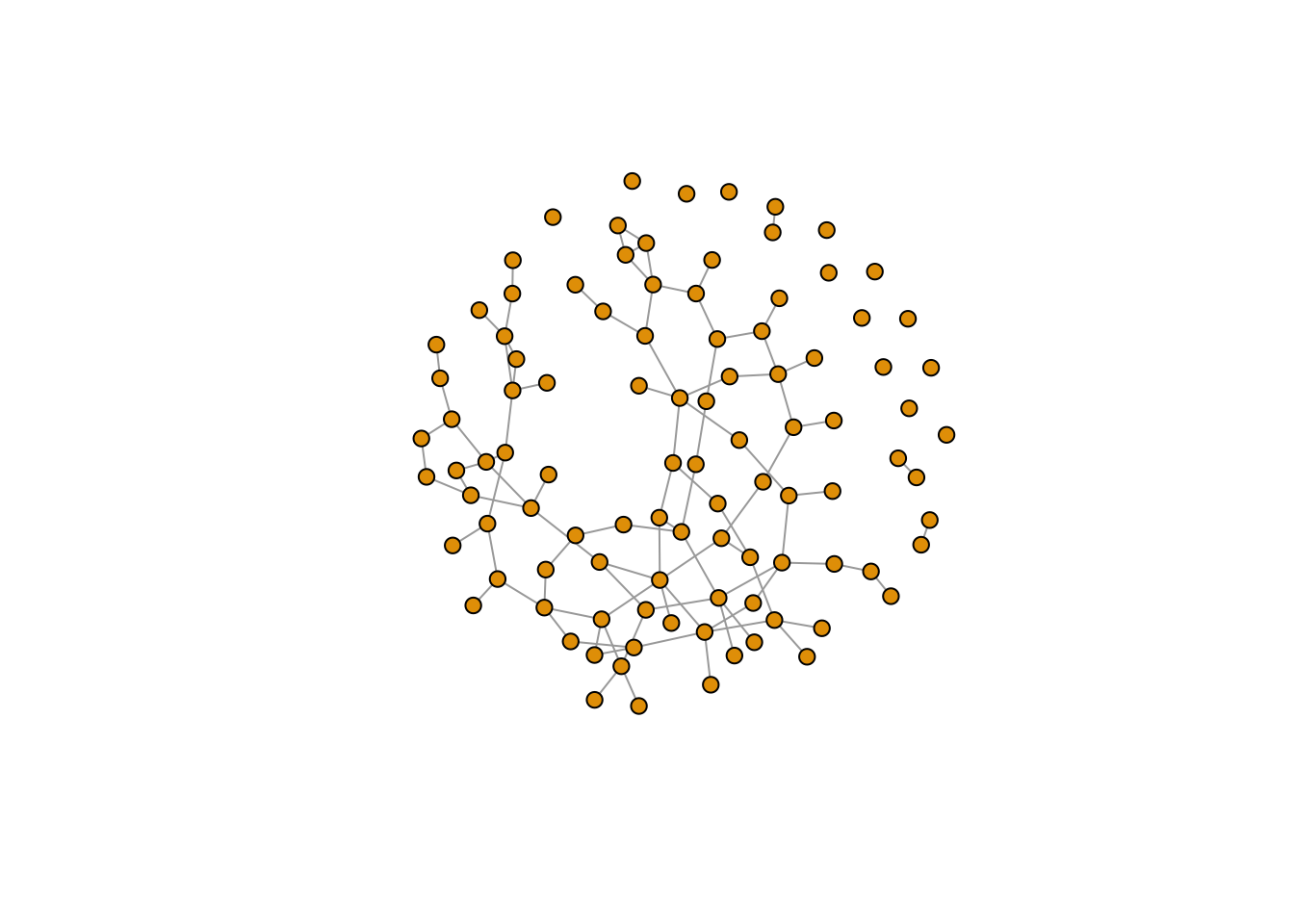
2.2.4 Adjustments on graphs
- igraph object as a layer (using
+) - igraph object as a matrix (using
[]) - rewiring a graph using
rewire,connect.neighborhood - combine graphs
%du% - other functions
2.2.4.1 igraph object as a layer
kite <- make_empty_graph(directed = FALSE) + vertices(LETTERS[1:10]) +
edges('A','B', 'B','D', 'C','D', 'D','E', 'E','G', 'F','G', 'G','H', 'H','I', 'I','J')
plot(kite)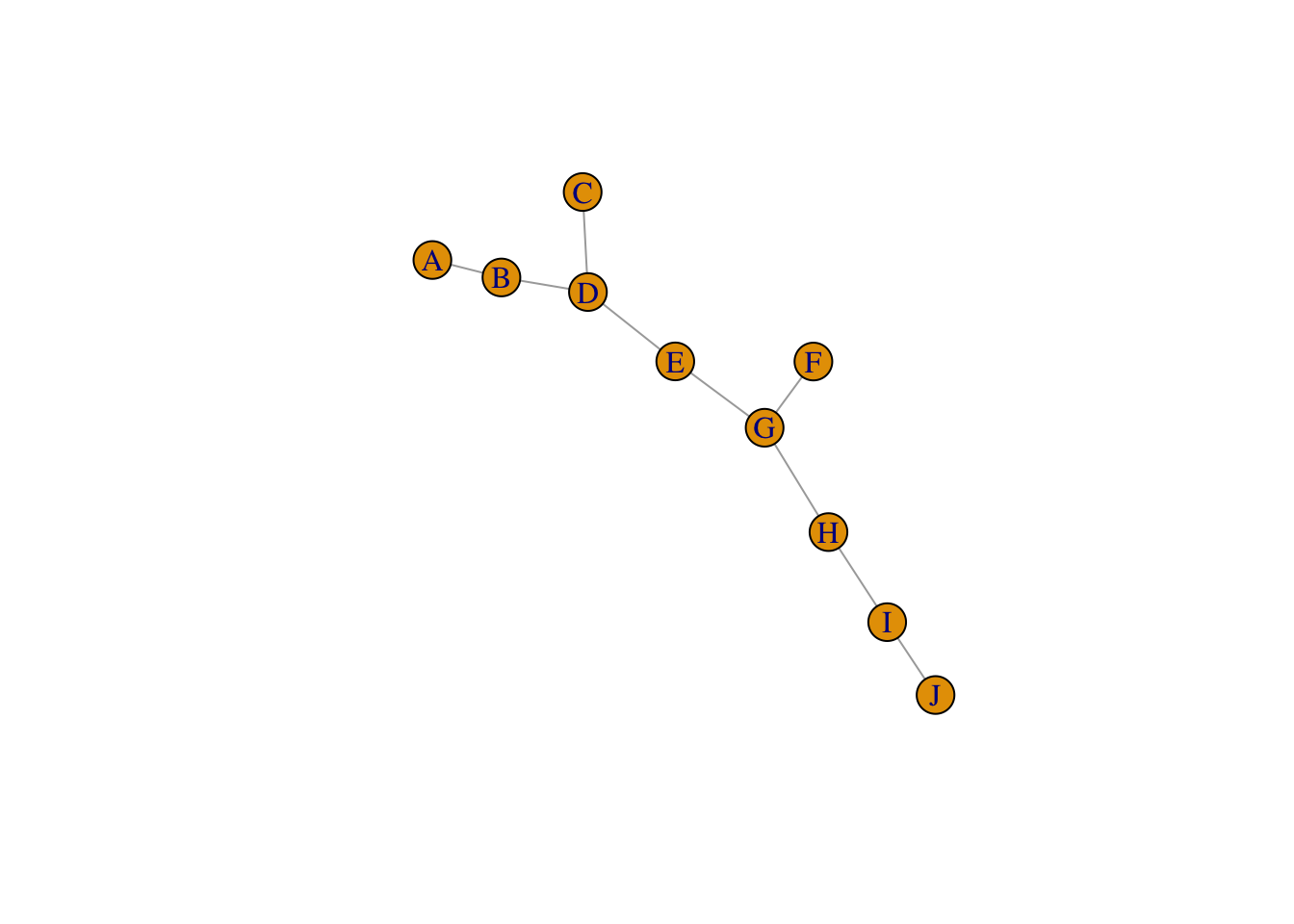
2.2.4.2 igraph object as a matrix
kite[]## 10 x 10 sparse Matrix of class "dgCMatrix"## [[ suppressing 10 column names 'A', 'B', 'C' ... ]]##
## A . 1 . . . . . . . .
## B 1 . . 1 . . . . . .
## C . . . 1 . . . . . .
## D . 1 1 . 1 . . . . .
## E . . . 1 . . 1 . . .
## F . . . . . . 1 . . .
## G . . . . 1 1 . 1 . .
## H . . . . . . 1 . 1 .
## I . . . . . . . 1 . 1
## J . . . . . . . . 1 .add edge
kite['A','F']=1
kite[]## 10 x 10 sparse Matrix of class "dgCMatrix"## [[ suppressing 10 column names 'A', 'B', 'C' ... ]]##
## A . 1 . . . 1 . . . .
## B 1 . . 1 . . . . . .
## C . . . 1 . . . . . .
## D . 1 1 . 1 . . . . .
## E . . . 1 . . 1 . . .
## F 1 . . . . . 1 . . .
## G . . . . 1 1 . 1 . .
## H . . . . . . 1 . 1 .
## I . . . . . . . 1 . 1
## J . . . . . . . . 1 .add multiple edges
kite[-1,1]## B C D E F G H I J
## 1 0 0 0 1 0 0 0 0kite[-1,1]=1
kite[] # add multiple edges or using from and to## 10 x 10 sparse Matrix of class "dgCMatrix"## [[ suppressing 10 column names 'A', 'B', 'C' ... ]]##
## A . 1 1 1 1 1 1 1 1 1
## B 1 . . 1 . . . . . .
## C 1 . . 1 . . . . . .
## D 1 1 1 . 1 . . . . .
## E 1 . . 1 . . 1 . . .
## F 1 . . . . . 1 . . .
## G 1 . . . 1 1 . 1 . .
## H 1 . . . . . 1 . 1 .
## I 1 . . . . . . 1 . 1
## J 1 . . . . . . . 1 .add multiple edges using from and to
kite[from=LETTERS[1:3],to=LETTERS[4:6]]=1
kite[]## 10 x 10 sparse Matrix of class "dgCMatrix"## [[ suppressing 10 column names 'A', 'B', 'C' ... ]]##
## A . 1 1 1 1 1 1 1 1 1
## B 1 . . 1 1 . . . . .
## C 1 . . 1 . 1 . . . .
## D 1 1 1 . 1 . . . . .
## E 1 1 . 1 . . 1 . . .
## F 1 . 1 . . . 1 . . .
## G 1 . . . 1 1 . 1 . .
## H 1 . . . . . 1 . 1 .
## I 1 . . . . . . 1 . 1
## J 1 . . . . . . . 1 .remove edge
kite[-1,2]=02.2.4.3 rewiring a graph
set.seed(1)
plot(rn, vertex.size=10, vertex.label=NA)
‘each_edge()’ is a rewiring method that changes the edge endpoints to a new vertex with a probability ‘prob’. And the new vertex is random variable distributed uniformly.
rn.rewired <- rewire(rn, each_edge(prob=0.1))
plot(rn.rewired, vertex.size=10, vertex.label=NA)
Rewire to connect vertices to other vertices at a certain distance.
rn.neigh = connect.neighborhood(rn, 5)
plot(rn.neigh, vertex.size=8, vertex.label=NA) 
g <- make_ring(10)
plot(g)
g <- connect(g, 2)
plot(g)
combine graphs
plot(rn %du% tr, vertex.size=10, vertex.label=NA)
2.2.5 Edge, vertex and network attributes
- Consider edge, vertex as sequences []
- Consider the network as matrix []
- Neighbors [[]]
- Attributes $
2.2.5.1 consider edge, vertex as sequences
plot(g4)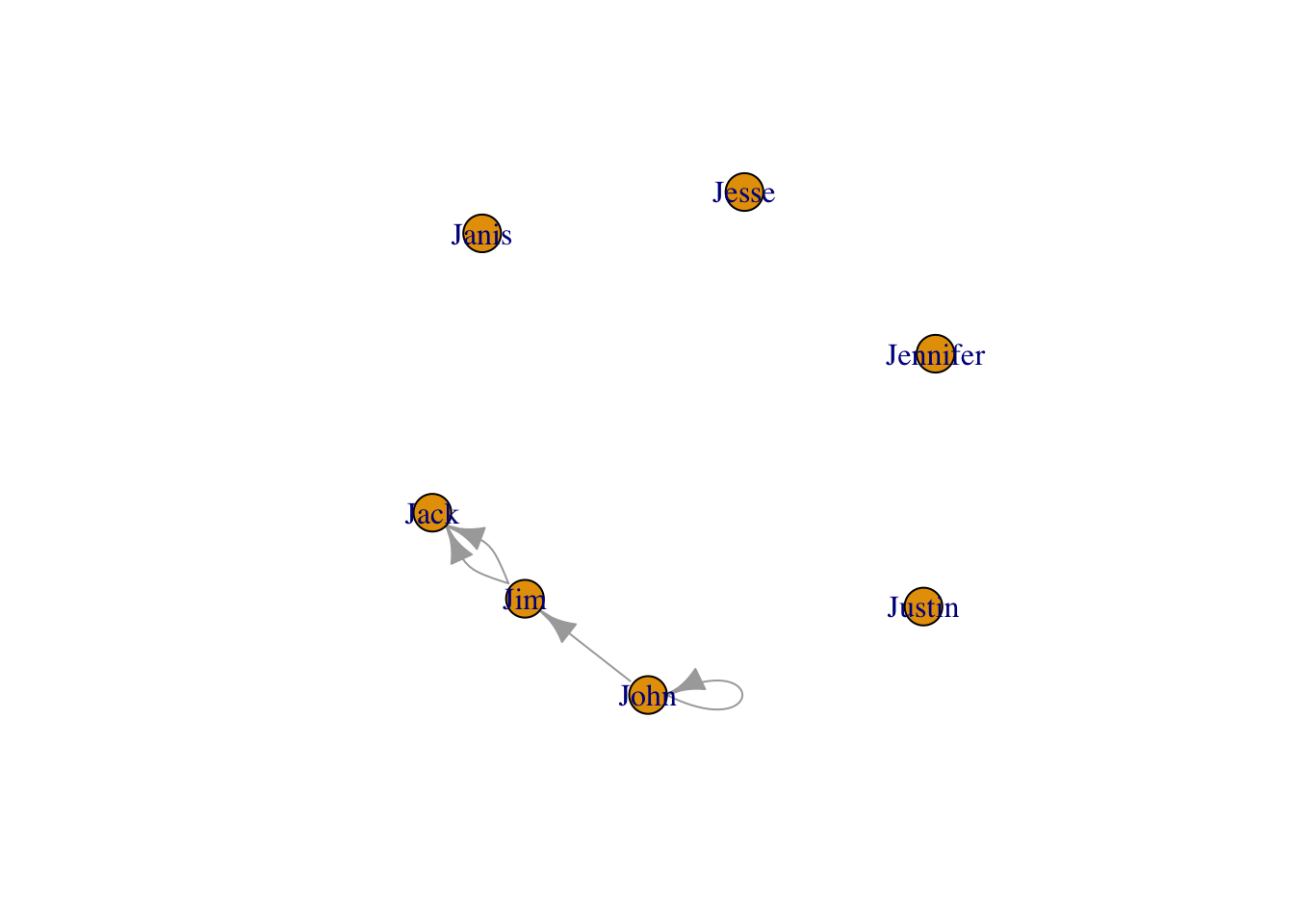
E(g4) #edge list## + 4/4 edges from 562dd0e (vertex names):
## [1] John->Jim Jim ->Jack Jim ->Jack John->JohnV(g4) #vertex list## + 7/7 vertices, named, from 562dd0e:
## [1] John Jim Jack Jesse Janis Jennifer Justinecount(g4) # count## [1] 4vcount(g4) # count## [1] 7refer vertex and edges
V(g4)[c("John","Jim")]## + 2/7 vertices, named, from 562dd0e:
## [1] John JimV(g4)[nei("Jim")] # neighbors of Jim## + 2/7 vertices, named, from 562dd0e:
## [1] John JackE(g4)[c("John|Jim","Jim|Jack")]## + 2/4 edges from 562dd0e (vertex names):
## [1] John->Jim Jim ->JackE(g4,path = c("John","Jim","Jack"))## + 2/4 edges from 562dd0e (vertex names):
## [1] John->Jim Jim ->JackE(g4)[ "John" %--% "Jack" ]## + 0/4 edges from 562dd0e (vertex names):E(g4)[ "Jim" %->% "Jack" ]## + 2/4 edges from 562dd0e (vertex names):
## [1] Jim->Jack Jim->JackE(g4)[ from("Jim") ]## + 2/4 edges from 562dd0e (vertex names):
## [1] Jim->Jack Jim->JackE(g4)[ to("Jim") ]## + 1/4 edge from 562dd0e (vertex names):
## [1] John->Jim2.2.5.2 consider the network as matrix
class(g4)## [1] "igraph"g4[] #"adjacency matrix"## 7 x 7 sparse Matrix of class "dgCMatrix"
## John Jim Jack Jesse Janis Jennifer Justin
## John 1 1 . . . . .
## Jim . . 2 . . . .
## Jack . . . . . . .
## Jesse . . . . . . .
## Janis . . . . . . .
## Jennifer . . . . . . .
## Justin . . . . . . .g4[1,] # consider as a matrix to select ## John Jim Jack Jesse Janis Jennifer Justin
## 1 1 0 0 0 0 0get.adjacency(g4) ## 7 x 7 sparse Matrix of class "dgCMatrix"
## John Jim Jack Jesse Janis Jennifer Justin
## John 1 1 . . . . .
## Jim . . 2 . . . .
## Jack . . . . . . .
## Jesse . . . . . . .
## Janis . . . . . . .
## Jennifer . . . . . . .
## Justin . . . . . . .##explicitly getting adjacency matrix (as a sparse matrix)
get.adjacency(g4,sparse=FALSE) ## John Jim Jack Jesse Janis Jennifer Justin
## John 1 1 0 0 0 0 0
## Jim 0 0 2 0 0 0 0
## Jack 0 0 0 0 0 0 0
## Jesse 0 0 0 0 0 0 0
## Janis 0 0 0 0 0 0 0
## Jennifer 0 0 0 0 0 0 0
## Justin 0 0 0 0 0 0 0##explicitly getting adjacency matrix --- not sparse, lets you manipulate it betterg4[1:2,2:3]## 2 x 2 sparse Matrix of class "dgCMatrix"
## Jim Jack
## John 1 .
## Jim . 2g4[from=c("Jack","Jim","John"),to=c("Jim","Jack","John")]## [1] 0 1 12.2.5.3 neighbors
neighbors(g4,"Jim")## + 2/7 vertices, named, from 562dd0e:
## [1] Jack Jackg4[["Jim"]]## $Jim
## + 2/7 vertices, named, from 562dd0e:
## [1] Jack Jackg4[[c("Jim","John")]] #works for multiple vertices## $Jim
## + 2/7 vertices, named, from 562dd0e:
## [1] Jack Jack
##
## $John
## + 2/7 vertices, named, from 562dd0e:
## [1] John Jimg4[["Jim",]]## $Jim
## + 2/7 vertices, named, from 562dd0e:
## [1] Jack Jackg4[[,"Jim"]]## $Jim
## + 1/7 vertex, named, from 562dd0e:
## [1] Johng4[[,"Jim",edges=TRUE]]## $Jim
## + 1/4 edge from 562dd0e (vertex names):
## [1] John->Jim2.2.5.4 Attributes: vertex attributes, edge attributes, graph attributes
use $ to create attributes and get attributes
V(g4)$name # automatically generated when we created the network.## [1] "John" "Jim" "Jack" "Jesse" "Janis" "Jennifer"
## [7] "Justin"V(g4)$gender <- c("male", "male", "male", "male", "female", "female", "male")
neighbors(g4,"Jim",mode = "all")$gender## [1] "male" "male" "male"E(g4)$type <- "email" # Edge attribute, assign "email" to all edges
E(g4)$weight <- 10 # Edge weight, setting all existing edges to 10
g4 <- set_graph_attr(g4, "name", "Email Network")see the attributes
edge_attr(g4)## $type
## [1] "email" "email" "email" "email"
##
## $weight
## [1] 10 10 10 10vertex_attr(g4)## $name
## [1] "John" "Jim" "Jack" "Jesse" "Janis" "Jennifer"
## [7] "Justin"
##
## $gender
## [1] "male" "male" "male" "male" "female" "female" "male"graph_attr(g4)## $name
## [1] "Email Network"graph_attr_names(g4)## [1] "name"graph_attr(g4, "name")## [1] "Email Network"can remove the attribute
g4 <- set_graph_attr(g4, "something", "A thing")
g4 <- delete_graph_attr(g4, "something")
graph_attr(g4)## $name
## [1] "Email Network"Make use of these attributes
plot(g4, edge.arrow.size=.5, vertex.label.color="black", vertex.label.dist=1.5,
vertex.color=as.factor(V(g4)$gender) )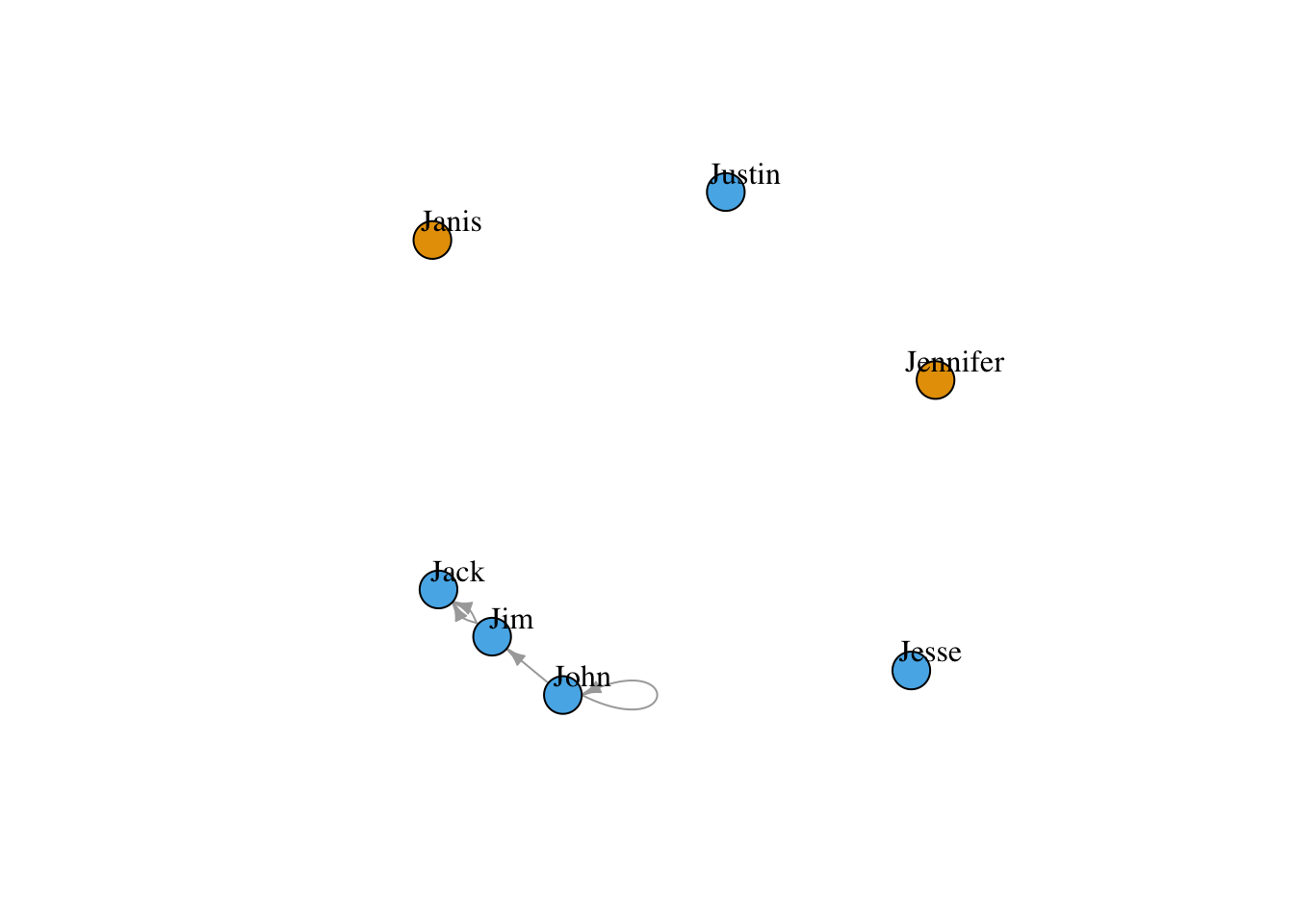
plot(g4, edge.arrow.size=.5, vertex.label.color="black", vertex.label.dist=1.5,
vertex.color=c( "pink", "skyblue")[1+(V(g4)$gender=="male")] ) 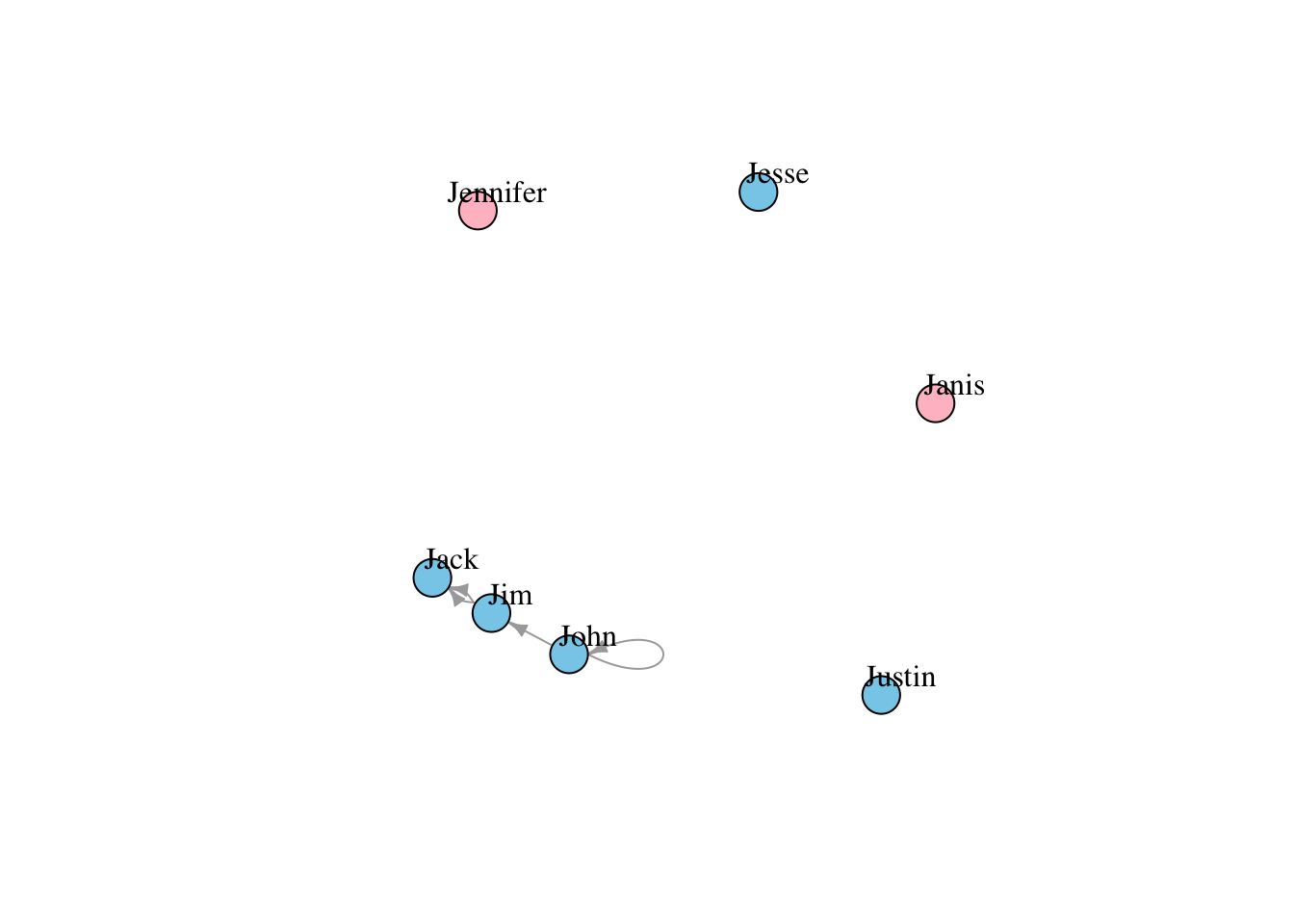
#consider as a sequenceattributes can be combined
plot(g4)
g4s <- igraph::simplify( g4, remove.multiple = T, remove.loops = F,
edge.attr.comb=c(weight="sum", type="ignore") )
#specifies what to do with edge attributes, if remove.multiple=TRUE. In this case many edges might be mapped to a single one in the new graph, and their attributes are combined.
E(g4)$type## [1] "email" "email" "email" "email"E(g4s)$type## NULLE(g4)$weight## [1] 10 10 10 10E(g4s)$weight## [1] 10 10 202.2.5.5 special attributes
make sure to avoid using these attribute names: color(e/v), layout(g), name(v),shape(v),type(v),weight(e)
2.2.6 Description of igraph object
g4s## IGRAPH 6cadc8c DNW- 7 3 -- Email Network
## + attr: name (g/c), name (v/c), gender (v/c), weight (e/n)
## + edges from 6cadc8c (vertex names):
## [1] John->John John->Jim Jim ->Jack- D or U, for a directed or undirected graph
- N for a named graph (where nodes have a name attribute)
- W for a weighted graph (where edges have a weight attribute)
- B for a bipartite (two-mode) graph (where nodes have a type attribute)
- (7 5) refer to the number of nodes and edges
- node & edge attributes, for example: g:graph; v: vertex; e: edge;n:numeric; c:character;l:logical; x:complex
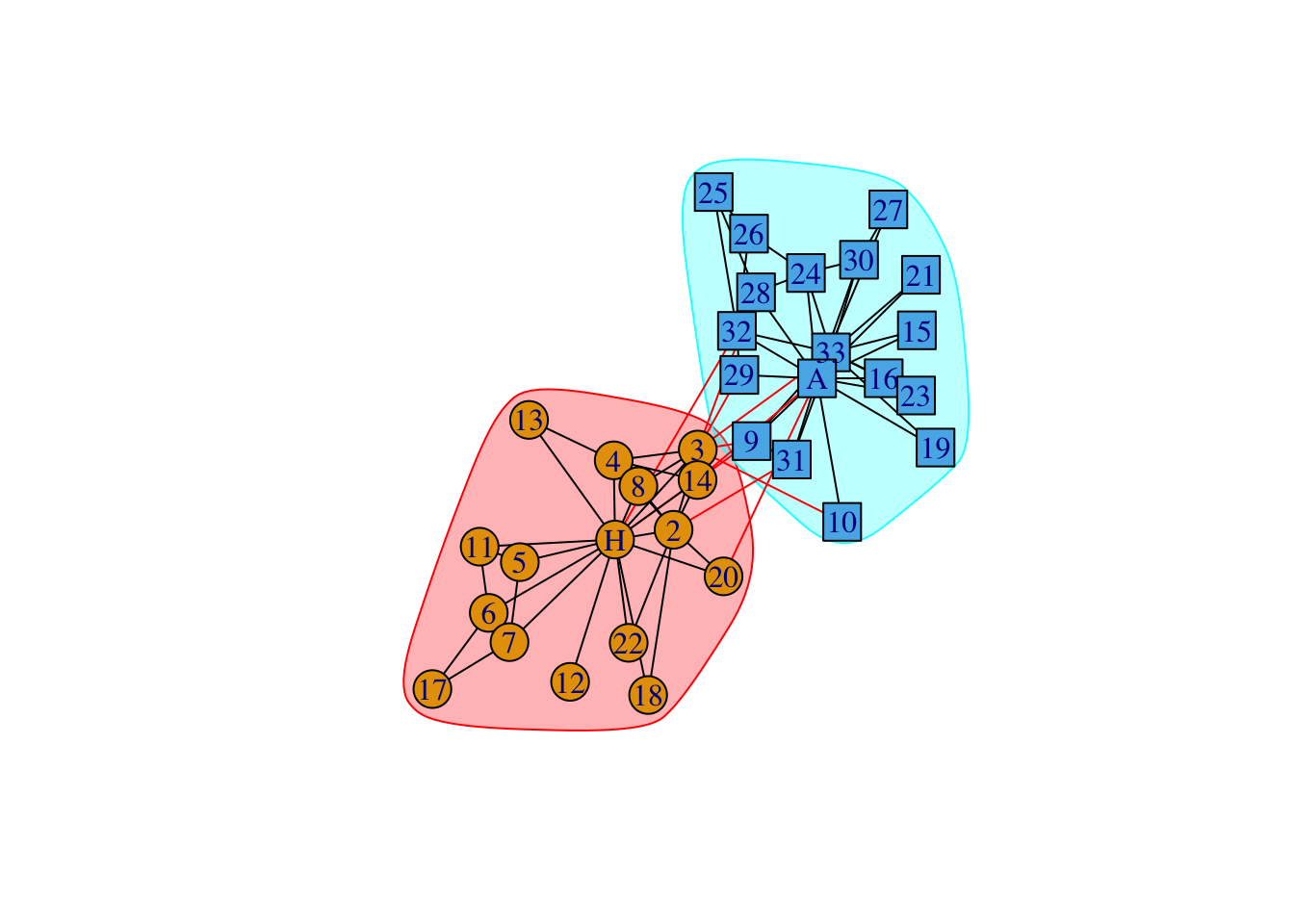
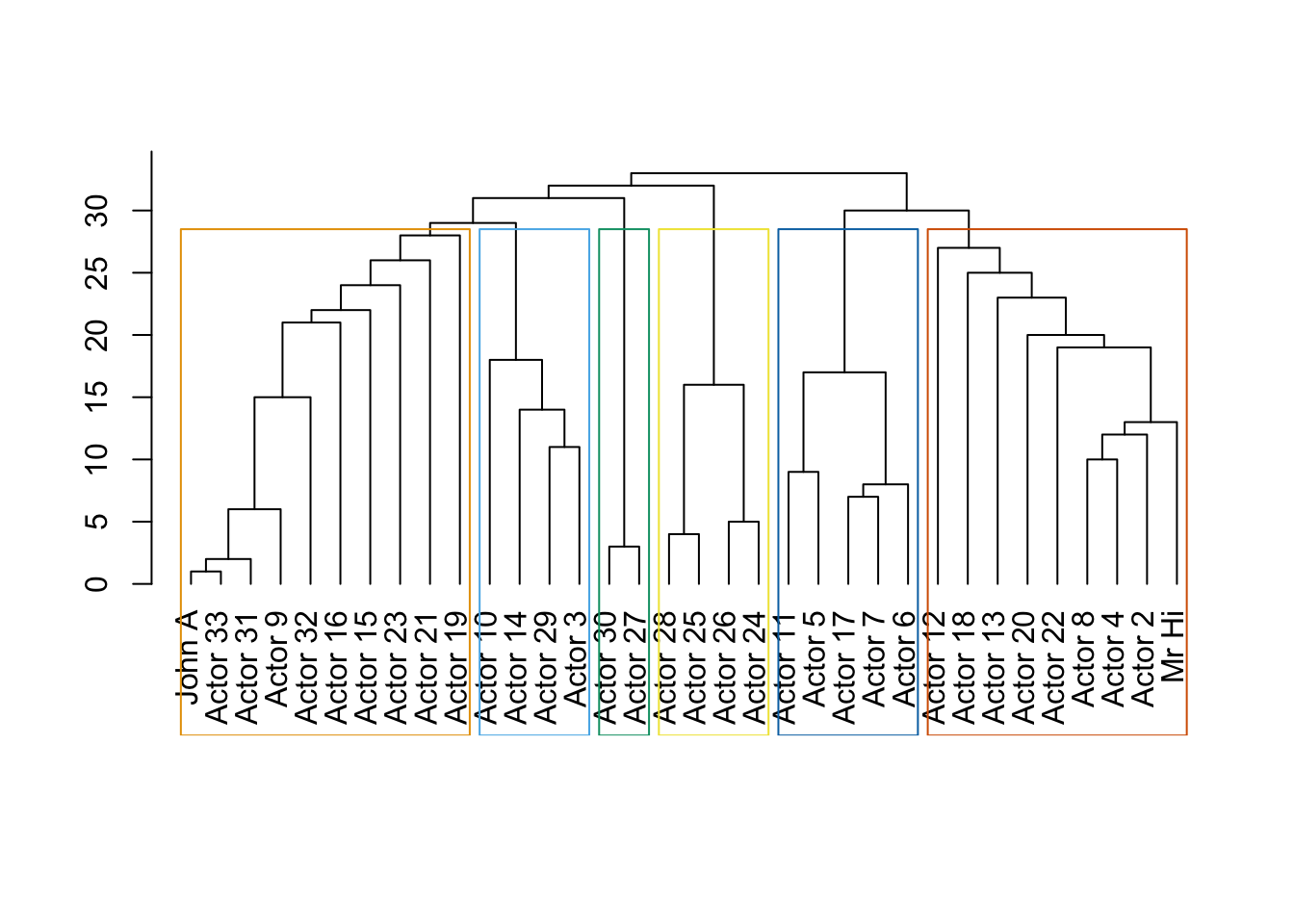
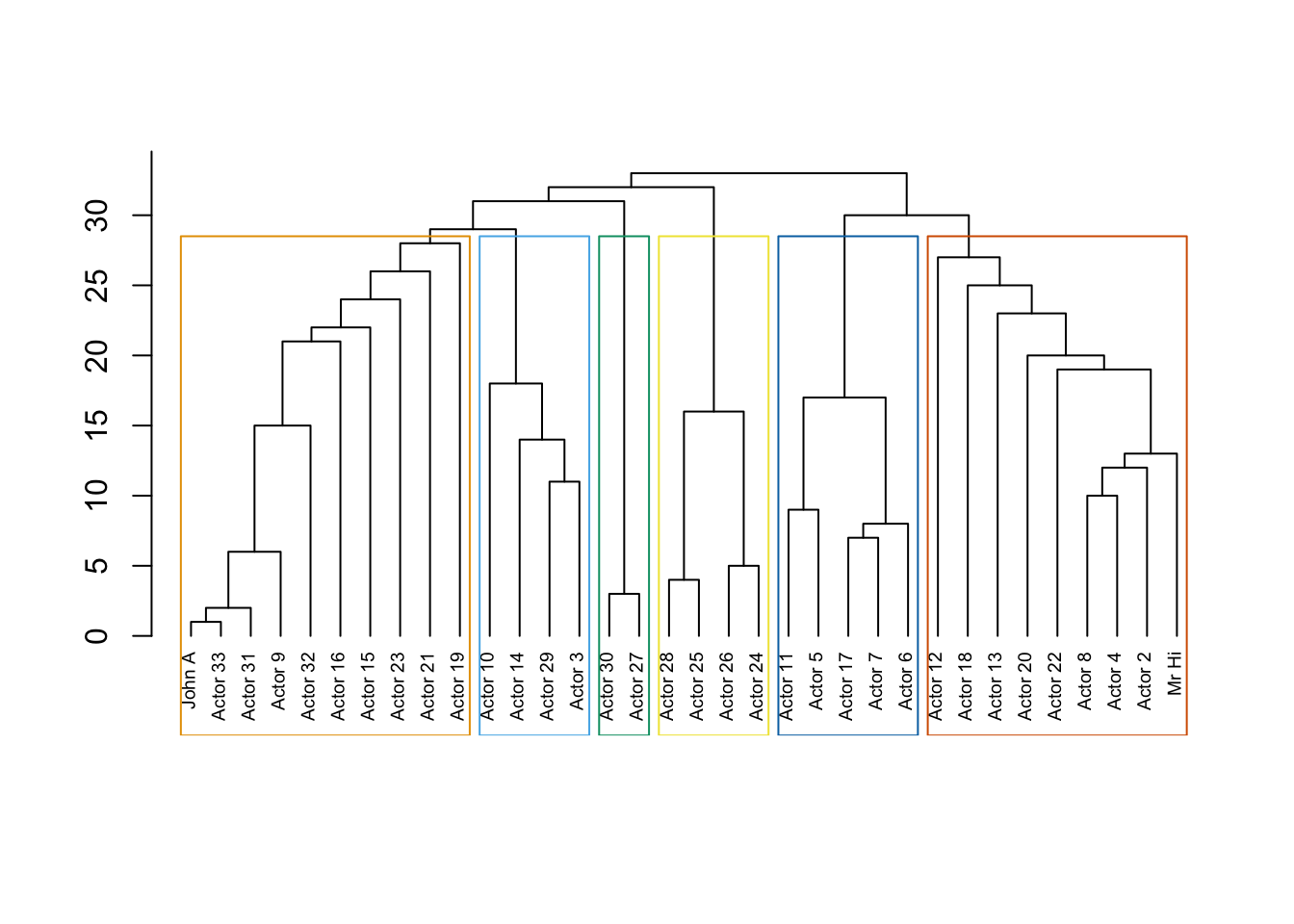
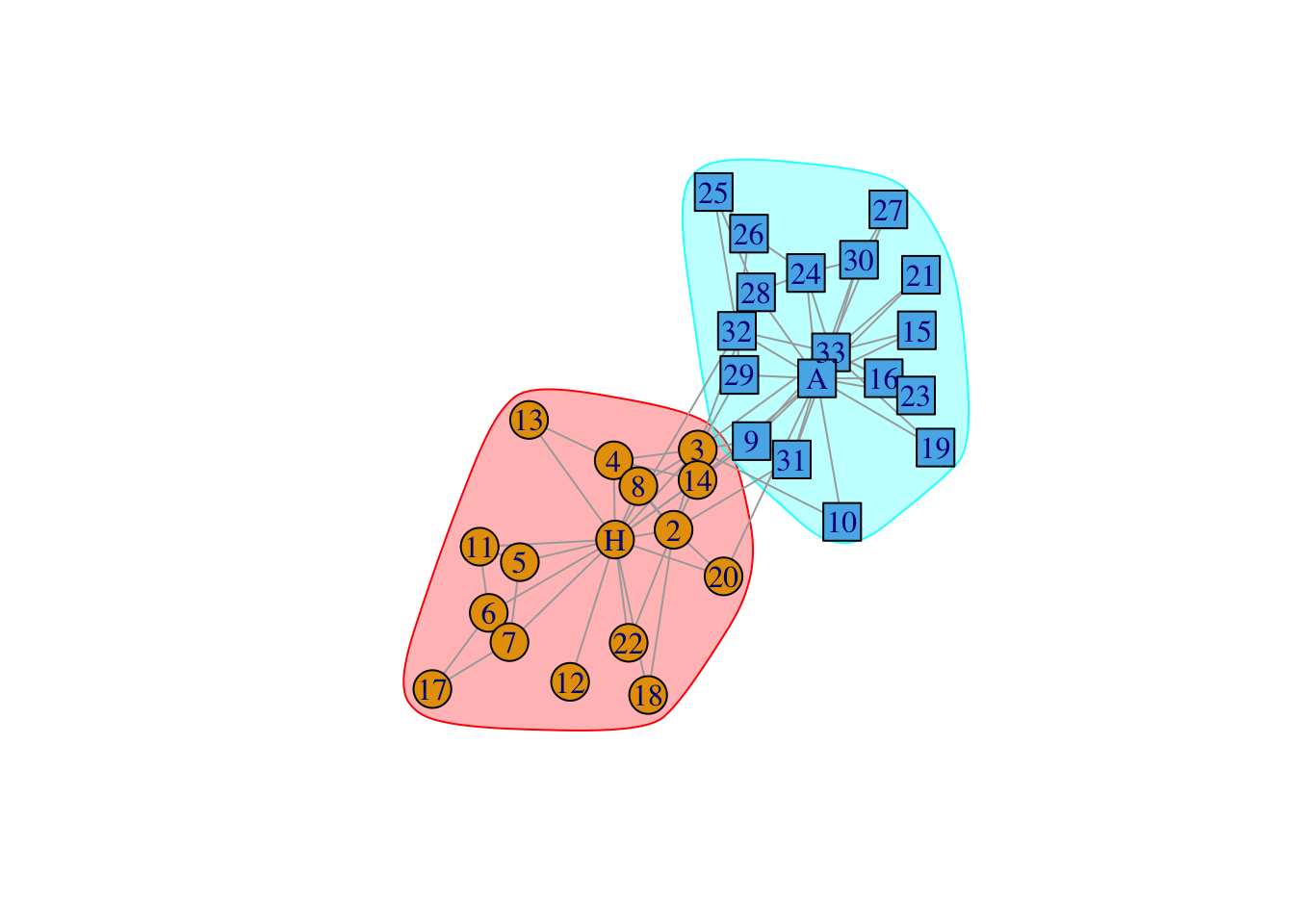
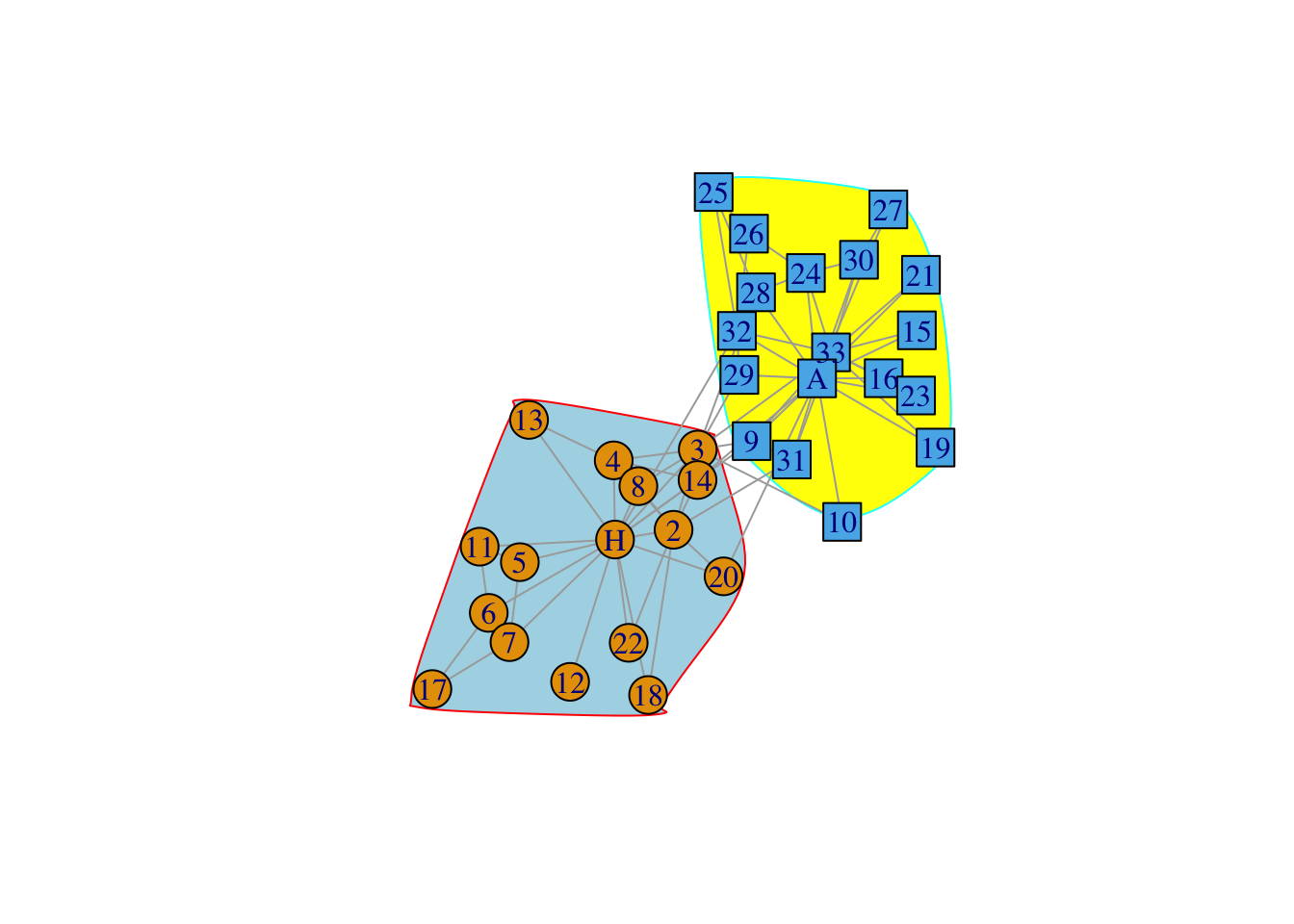
data(karate)
karate## IGRAPH 4b458a1 UNW- 34 78 -- Zachary's karate club network
## + attr: name (g/c), Citation (g/c), Author (g/c), Faction (v/n),
## | name (v/c), label (v/c), color (v/n), weight (e/n)
## + edges from 4b458a1 (vertex names):
## [1] Mr Hi --Actor 2 Mr Hi --Actor 3 Mr Hi --Actor 4
## [4] Mr Hi --Actor 5 Mr Hi --Actor 6 Mr Hi --Actor 7
## [7] Mr Hi --Actor 8 Mr Hi --Actor 9 Mr Hi --Actor 11
## [10] Mr Hi --Actor 12 Mr Hi --Actor 13 Mr Hi --Actor 14
## [13] Mr Hi --Actor 18 Mr Hi --Actor 20 Mr Hi --Actor 22
## [16] Mr Hi --Actor 32 Actor 2--Actor 3 Actor 2--Actor 4
## [19] Actor 2--Actor 8 Actor 2--Actor 14 Actor 2--Actor 18
## + ... omitted several edgesdata(macaque)
macaque## IGRAPH f7130f3 DN-- 45 463 --
## + attr: Citation (g/c), Author (g/c), shape (v/c), name (v/c)
## + edges from f7130f3 (vertex names):
## [1] V1 ->V2 V1 ->V3 V1 ->V3A V1 ->V4 V1 ->V4t
## [6] V1 ->MT V1 ->PO V1 ->PIP V2 ->V1 V2 ->V3
## [11] V2 ->V3A V2 ->V4 V2 ->V4t V2 ->VOT V2 ->VP
## [16] V2 ->MT V2 ->MSTd/p V2 ->MSTl V2 ->PO V2 ->PIP
## [21] V2 ->VIP V2 ->FST V2 ->FEF V3 ->V1 V3 ->V2
## [26] V3 ->V3A V3 ->V4 V3 ->V4t V3 ->MT V3 ->MSTd/p
## [31] V3 ->PO V3 ->LIP V3 ->PIP V3 ->VIP V3 ->FST
## [36] V3 ->TF V3 ->FEF V3A->V1 V3A->V2 V3A->V3
## + ... omitted several edges2.3 Built networks from external sources, basic visualization and more on network descriptions
2.3.1 Outline
- Get network from files (edgelist, matrix, dataframe)
- Visualization
- Plotting parameters
- Layouts
- Network and node descriptions
2.3.2 Dataset
- Datasets: Download the data from my github.
- The full dataset comes from https://github.com/mathbeveridge/asoiaf
- Analysis on the datasets: https://www.macalester.edu/~abeverid/thrones.html
](images/got-network.png)
Figure 2.2: Network Visualization from abeverid
2.3.3 Get network from files
2.3.3.1 Creating network
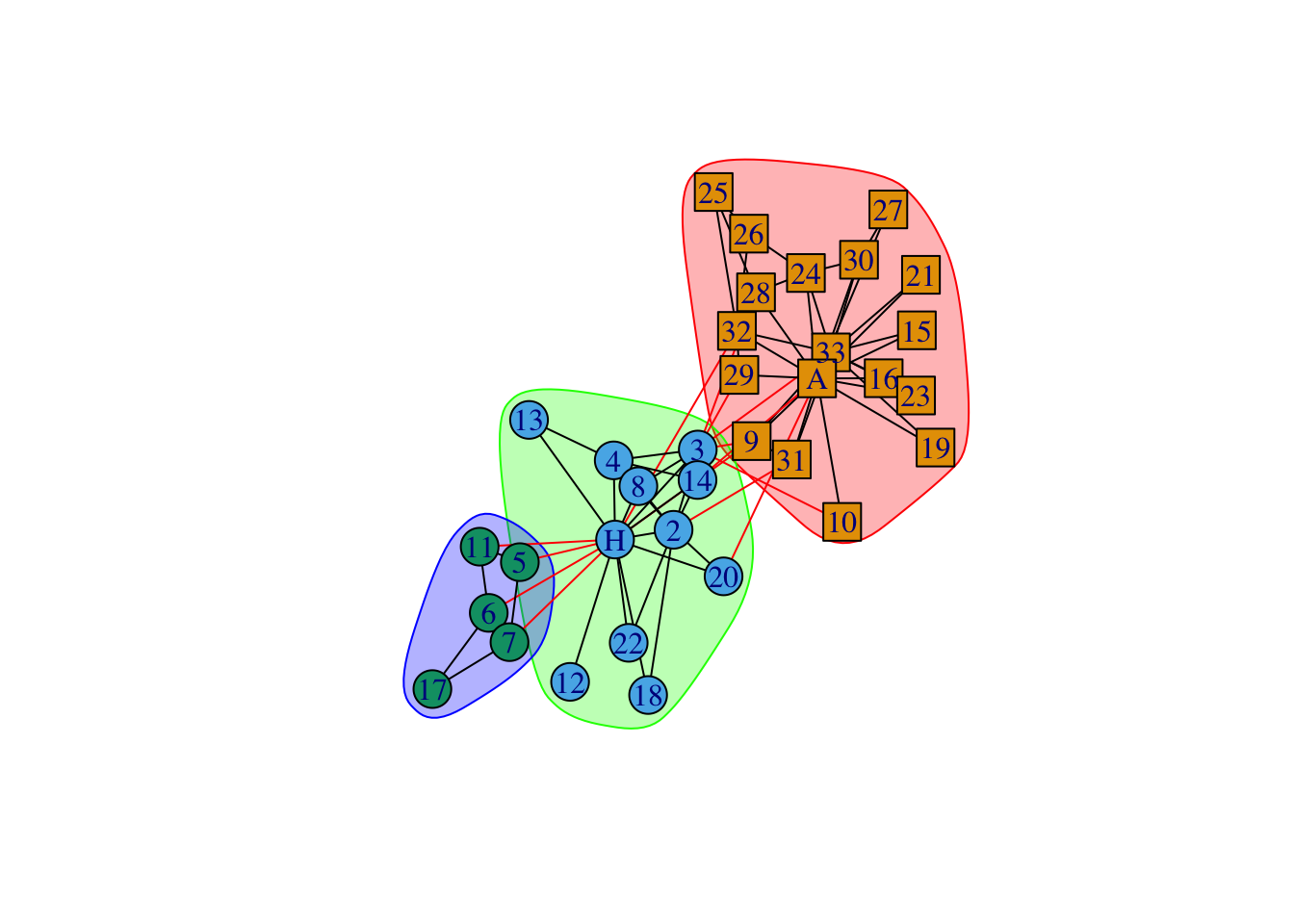
](images/cnet_all.png)
Figure 2.3: Introduction from igraph manual
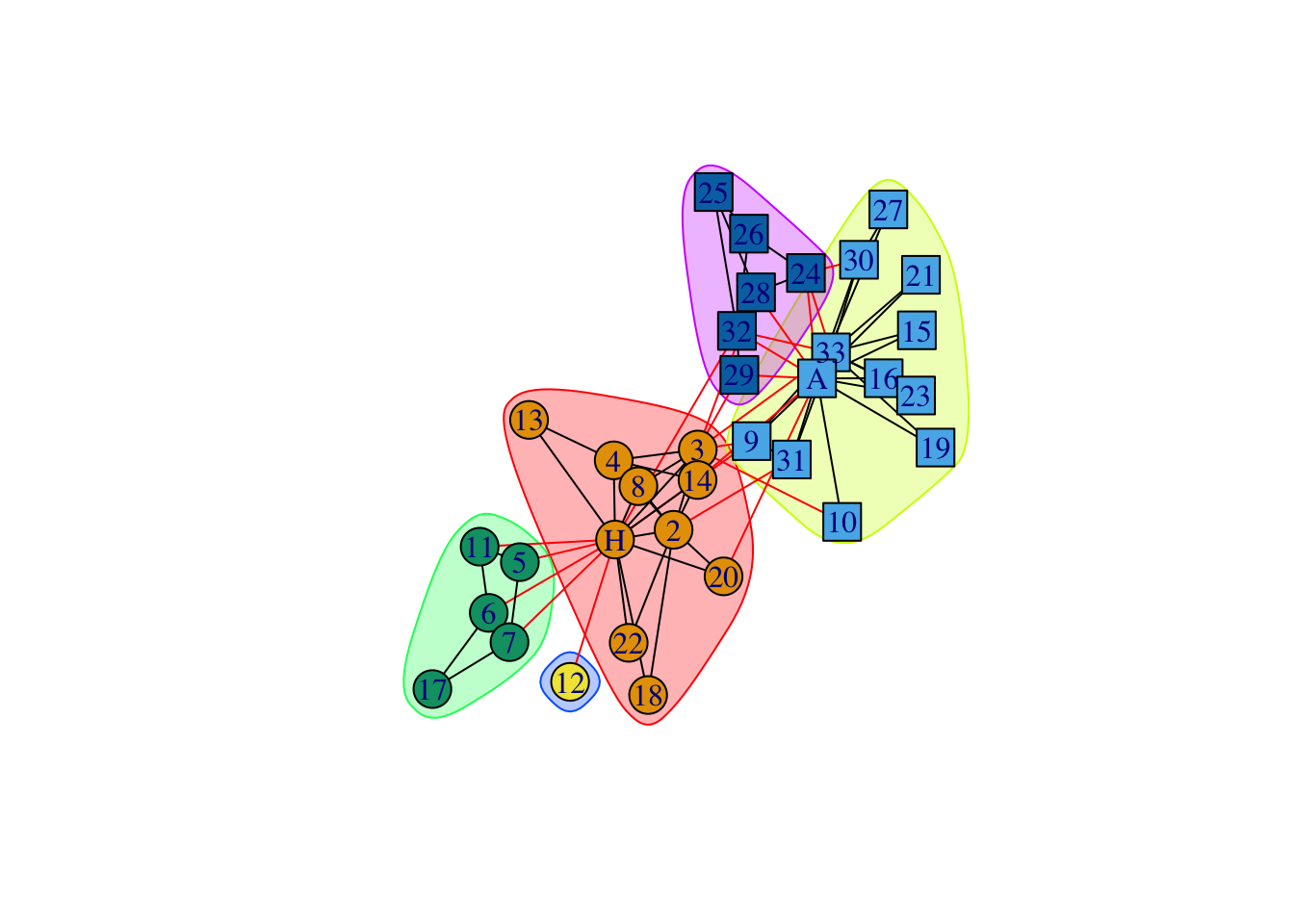
](images/cnet1.png)
Figure 2.4: Introduction from igraph manual
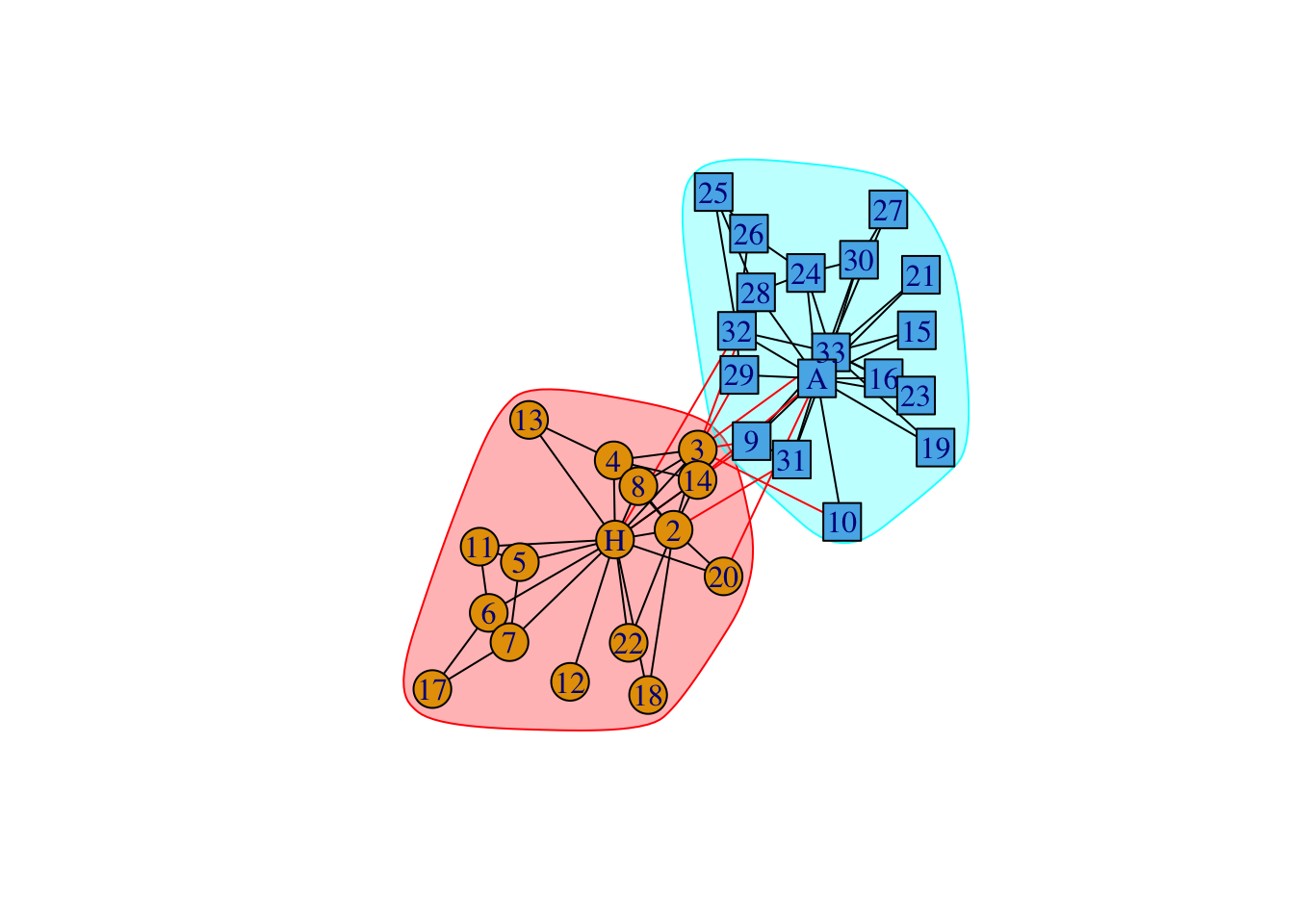
](images/cnet3.png)
Figure 2.5: Introduction from igraph manual
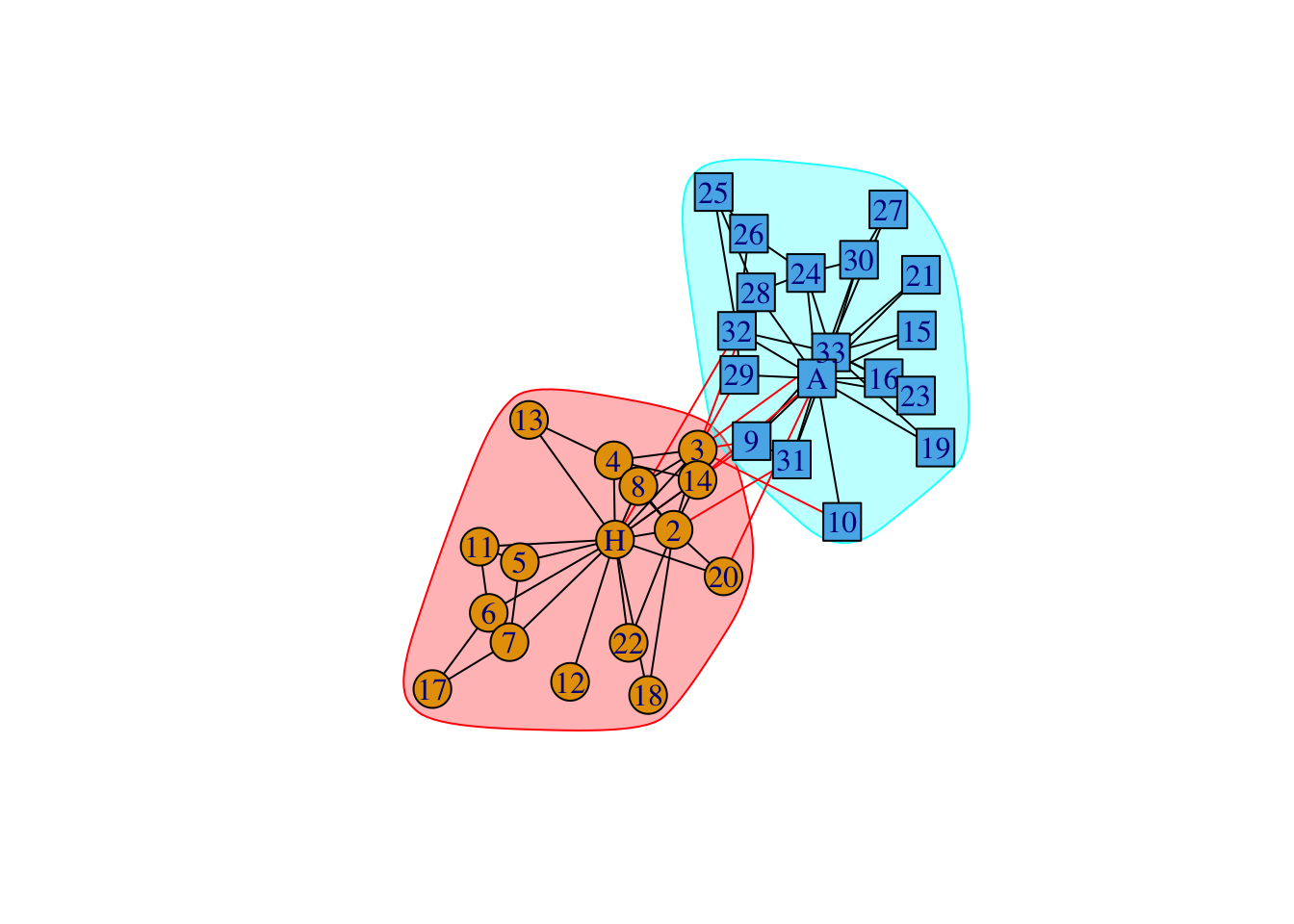
](images/cnet2.png)
Figure 2.6: Introduction from igraph manual
](images/cnet4.png)
Figure 2.7: Introduction from igraph manual
2.3.3.2 Get network from files
graph_from_adjacency_matrix()graph_from_edgelist()graph_from_data_frame()
2.3.3.3 graph_from_adjacency_matrix()
Used for creating a small matrix.
The networks in real world are usually large sparse matrix and stored as a edgelist.
Binary matrix:
set.seed(2)
#sample from Bernoulli distribution with sample size 100.
adjm <- matrix(sample(0:1, 100, replace=TRUE, prob=c(0.9,0.1)), nc=10)
adjm## [,1] [,2] [,3] [,4] [,5] [,6] [,7] [,8] [,9] [,10]
## [1,] 0 0 0 0 1 0 0 0 0 1
## [2,] 0 0 0 0 0 0 0 0 0 0
## [3,] 0 0 0 0 0 0 0 0 0 0
## [4,] 0 0 0 0 0 1 0 0 0 0
## [5,] 1 0 0 0 1 0 0 0 0 0
## [6,] 1 0 0 0 0 0 0 0 0 0
## [7,] 0 1 0 0 1 0 0 0 1 0
## [8,] 0 0 0 0 0 1 0 0 0 0
## [9,] 0 0 1 0 0 0 0 0 0 0
## [10,] 0 0 0 0 0 0 0 0 0 0g1 <- graph_from_adjacency_matrix( adjm )
set.seed(1)
plot(g1)
#default is directed
g2 <- graph_from_adjacency_matrix( adjm ,mode = "undirected")
set.seed(1)
plot(g2)
#get rid of the self-loop (in real-world maybe self-loop does not make any sense)
g3 <- graph_from_adjacency_matrix( adjm ,mode = "undirected",diag = FALSE)
set.seed(1)
plot(g3)
Sparse matrix:
adjms=g1[]
adjms## 10 x 10 sparse Matrix of class "dgCMatrix"
##
## [1,] . . . . 1 . . . . 1
## [2,] . . . . . . . . . .
## [3,] . . . . . . . . . .
## [4,] . . . . . 1 . . . .
## [5,] 1 . . . 1 . . . . .
## [6,] 1 . . . . . . . . .
## [7,] . 1 . . 1 . . . 1 .
## [8,] . . . . . 1 . . . .
## [9,] . . 1 . . . . . . .
## [10,] . . . . . . . . . .g4=graph_from_adjacency_matrix(adjms)
set.seed(1)
plot(g4)
Weighted matrix
set.seed(1)
adjmw <- matrix(sample(0:5, 100, replace=TRUE,
prob=c(0.9,0.02,0.02,0.02,0.02,0.02)), nc=10)
adjmw## [,1] [,2] [,3] [,4] [,5] [,6] [,7] [,8] [,9] [,10]
## [1,] 0 0 3 0 0 0 2 0 0 0
## [2,] 0 0 0 0 0 0 0 0 0 0
## [3,] 0 0 0 0 0 0 0 0 0 0
## [4,] 2 0 0 0 0 0 0 0 0 0
## [5,] 0 0 0 0 0 0 0 0 0 0
## [6,] 0 0 0 0 0 0 0 0 0 0
## [7,] 4 0 0 0 0 0 0 0 0 0
## [8,] 0 1 0 0 0 0 0 0 0 0
## [9,] 0 0 0 0 0 0 0 0 0 0
## [10,] 0 0 0 0 0 0 0 5 0 0g5 <- graph_from_adjacency_matrix(adjmw, weighted=TRUE)
set.seed(1)
plot(g5)
g5## IGRAPH 6f0feb3 D-W- 10 6 --
## + attr: weight (e/n)
## + edges from 6f0feb3:
## [1] 1->3 1->7 4->1 7->1 8->2 10->8E(g5)$weight## [1] 3 2 2 4 1 5Named matrix
rownames(adjmw)=LETTERS[1:10]
colnames(adjmw)=LETTERS[1:10]
g6 <- graph_from_adjacency_matrix(adjmw, weighted=TRUE)
set.seed(1)
plot(g6)
2.3.3.4 graph_from_edgelist()
Most network datasets are stored as edgelists. Input is two-column matrix with each row defining one edge.
gotdf=read.csv("images/gotstark_lannister.csv",stringsAsFactors = FALSE)
head(gotdf,5)## X Source Target Type weight book source.family
## 1 1 Arya-Stark Benjen-Stark Undirected 3 1 Stark
## 2 2 Arya-Stark Bran-Stark Undirected 14 1 Stark
## 3 3 Arya-Stark Catelyn-Stark Undirected 5 1 Stark
## 4 4 Arya-Stark Cersei-Lannister Undirected 12 1 Stark
## 5 5 Arya-Stark Desmond Undirected 3 1 Stark
## target.family
## 1 Stark
## 2 Stark
## 3 Stark
## 4 Lannister
## 5 <NA>library(dplyr)
library(tidyr)gotdf.el=gotdf%>%select(Source,Target,weight)%>%
group_by(Source,Target)%>%
expand(edge=c(1:weight))%>%select(-edge)
head(gotdf.el)## # A tibble: 6 x 2
## # Groups: Source, Target [2]
## Source Target
## <chr> <chr>
## 1 Arya-Stark Benjen-Stark
## 2 Arya-Stark Benjen-Stark
## 3 Arya-Stark Benjen-Stark
## 4 Arya-Stark Bran-Stark
## 5 Arya-Stark Bran-Stark
## 6 Arya-Stark Bran-Stark## input need to be a matrix
got1=graph_from_edgelist(gotdf.el%>%as.matrix(),directed = FALSE)
got1## IGRAPH bdd718b UN-- 99 3374 --
## + attr: name (v/c)
## + edges from bdd718b (vertex names):
## [1] Arya-Stark--Benjen-Stark Arya-Stark--Benjen-Stark
## [3] Arya-Stark--Benjen-Stark Arya-Stark--Bran-Stark
## [5] Arya-Stark--Bran-Stark Arya-Stark--Bran-Stark
## [7] Arya-Stark--Bran-Stark Arya-Stark--Bran-Stark
## [9] Arya-Stark--Bran-Stark Arya-Stark--Bran-Stark
## [11] Arya-Stark--Bran-Stark Arya-Stark--Bran-Stark
## [13] Arya-Stark--Bran-Stark Arya-Stark--Bran-Stark
## [15] Arya-Stark--Bran-Stark Arya-Stark--Bran-Stark
## + ... omitted several edgesplot(got1,edge.arrow.size=.5, vertex.color="gold", vertex.size=3,
vertex.frame.color="gray", vertex.label.color="black",
vertex.label.cex=.5, vertex.label.dist=2, edge.curved=0.2)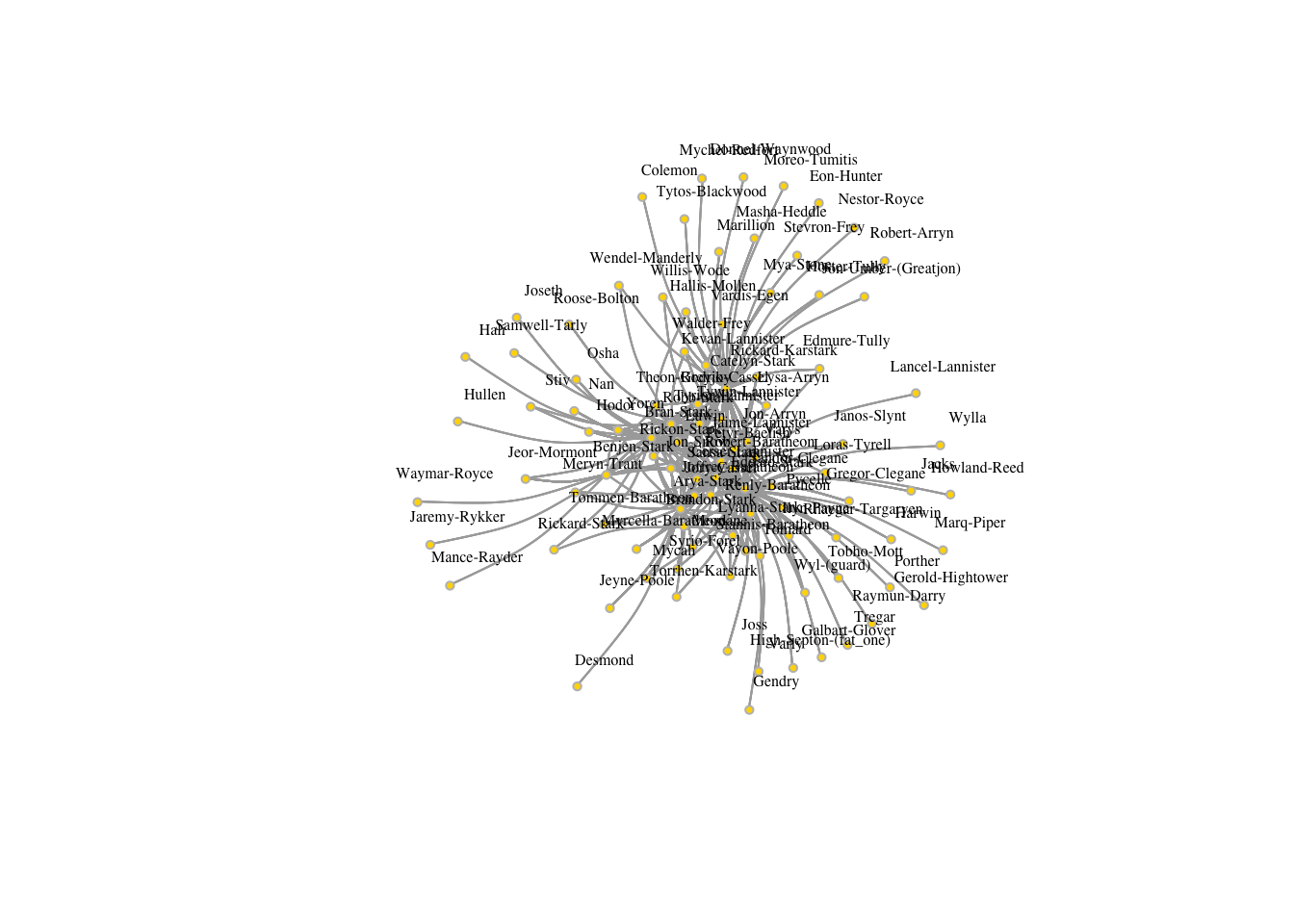
2.3.3.4.1 Simplify the network
el <- matrix( c("foo", "bar","foo","bar", "bar", "foobar"), nc = 2, byrow = TRUE)
graph_from_edgelist(el)%>%plot()
E(got1)$weight=rep(1,ecount(got1))
got1s <- igraph::simplify( got1, remove.multiple = T, remove.loops = F,
edge.attr.comb=c(weight="sum"))
plot(got1s,edge.arrow.size=.5, vertex.color="gold", vertex.size=3,
vertex.frame.color="gray", vertex.label.color="black",
vertex.label.cex=.5, vertex.label.dist=2, edge.curved=0.5,layout=layout_with_lgl)
2.3.3.4.2 Short name
library(stringr)nameshort=V(got1s)$name%>%
str_split(.,"-",simplify = TRUE)%>%
.[,1]
V(got1s)$name[1:3]## [1] "Arya-Stark" "Benjen-Stark" "Bran-Stark"nameshort[1:3]## [1] "Arya" "Benjen" "Bran"V(got1s)$name=nameshort
plot(got1s,edge.arrow.size=.5, vertex.color="gold", vertex.size=3,
vertex.frame.color="gray", vertex.label.color="black",
vertex.label.cex=.5, vertex.label.dist=2, edge.curved=0.5,layout=layout_with_lgl)
2.3.3.5 graph_from_data_frame()
Most common and useful.
d: a data frame containing a symbolic edge list in the first two columns. Additional columns are considered as edge attributes.
vertices: A data frame with vertex metadata
head(gotdf,5)## X Source Target Type weight book source.family
## 1 1 Arya-Stark Benjen-Stark Undirected 3 1 Stark
## 2 2 Arya-Stark Bran-Stark Undirected 14 1 Stark
## 3 3 Arya-Stark Catelyn-Stark Undirected 5 1 Stark
## 4 4 Arya-Stark Cersei-Lannister Undirected 12 1 Stark
## 5 5 Arya-Stark Desmond Undirected 3 1 Stark
## target.family
## 1 Stark
## 2 Stark
## 3 Stark
## 4 Lannister
## 5 <NA>gotdf=gotdf%>%select(-X)
got2=graph_from_data_frame(d=gotdf,directed = FALSE)
got2## IGRAPH 21f2988 UNW- 99 238 --
## + attr: name (v/c), Type (e/c), weight (e/n), book (e/n),
## | source.family (e/c), target.family (e/c)
## + edges from 21f2988 (vertex names):
## [1] Arya-Stark--Benjen-Stark Arya-Stark--Bran-Stark
## [3] Arya-Stark--Catelyn-Stark Arya-Stark--Cersei-Lannister
## [5] Arya-Stark--Desmond Arya-Stark--Eddard-Stark
## [7] Arya-Stark--Ilyn-Payne Arya-Stark--Jeyne-Poole
## [9] Arya-Stark--Joffrey-Baratheon Arya-Stark--Jon-Snow
## [11] Arya-Stark--Jory-Cassel Arya-Stark--Meryn-Trant
## [13] Arya-Stark--Mordane Arya-Stark--Mycah
## + ... omitted several edgesplot(got2,edge.arrow.size=.5, vertex.color="gold", vertex.size=3,
vertex.frame.color="gray", vertex.label.color="black",
vertex.label.cex=.5, vertex.label.dist=2, edge.curved=0.5,layout=layout_with_lgl)
2.3.3.5.1 get dataframe, matrix or adgelist from igraph object
igraph::as_data_frame(got2)%>%head(2)## from to Type weight book source.family
## 1 Arya-Stark Benjen-Stark Undirected 3 1 Stark
## 2 Arya-Stark Bran-Stark Undirected 14 1 Stark
## target.family
## 1 Stark
## 2 Starkas_adjacency_matrix(got2)%>%head(2)## [1] 0 1as_edgelist(got2)%>%head(2)## [,1] [,2]
## [1,] "Arya-Stark" "Benjen-Stark"
## [2,] "Arya-Stark" "Bran-Stark"2.3.3.5.2 read_graph, write_graph
## store in txt or csv or others
write_graph(graph = got2,file = "g.txt",format = "edgelist")
read_graph(file = "g.txt",format = "edgelist",directed=F)## IGRAPH 99ad3df U--- 99 238 --
## + edges from 99ad3df:
## [1] 1-- 2 1-- 3 1-- 5 1-- 6 1-- 7 1--12 1--13 1--14 1--17 1--18 1--19
## [12] 1--20 1--21 1--22 1--23 1--24 1--25 1--26 1--27 1--28 1--29 1--30
## [23] 1--31 1--32 1--33 1--34 1--35 2-- 3 2-- 6 2--13 2--15 2--21 2--28
## [34] 2--35 2--36 2--37 2--38 2--39 2--40 2--41 3-- 5 3-- 6 3-- 7 3--12
## [45] 3--13 3--14 3--15 3--20 3--21 3--22 3--27 3--28 3--29 3--33 3--35
## [56] 3--37 3--38 3--40 3--42 3--43 3--44 3--45 3--46 3--47 3--48 3--49
## [67] 3--50 3--51 3--52 3--53 4-- 7 4--11 4--27 4--28 4--52 5-- 6 5-- 7
## [78] 5-- 8 5--12 5--13 5--14 5--15 5--16 5--20 5--21 5--27 5--28 5--29
## [89] 5--38 5--40 5--43 5--46 5--51 5--54 5--55 5--56 5--57 5--58 5--59
## + ... omitted several edges## store the whole graph
write_graph(got2,file = "gg",format = "pajek")
read_graph(file="gg",format="pajek")## IGRAPH 6586ad6 U-W- 99 238 --
## + attr: weight (e/n)
## + edges from 6586ad6:
## [1] 1-- 2 1-- 3 1-- 5 1-- 6 1--17 1-- 7 1--18 1--19 1--20 1--21 1--22
## [12] 1--23 1--24 1--25 1--26 1--27 1--12 1--13 1--28 1--29 1--30 1--14
## [23] 1--31 1--32 1--33 1--34 1--35 2-- 3 2-- 6 2--36 2--37 2--21 2--38
## [34] 2--39 2--13 2--28 2--40 2--15 2--41 2--35 3-- 5 3-- 6 3-- 7 3--42
## [45] 3--43 3--44 3--45 3--37 3--20 3--46 3--21 3--22 3--47 3--38 3--48
## [56] 3--49 3--27 3--50 3--51 3--52 3--12 3--13 3--28 3--29 3--14 3--53
## [67] 3--40 3--33 3--15 3--35 4-- 7 4--11 4--27 4--52 4--28 5-- 6 5--54
## [78] 5--55 5-- 7 5--56 5--57 5--43 5--58 5-- 8 5--20 5--46 5--21 5--59
## + ... omitted several edgesgot2## IGRAPH 21f2988 UNW- 99 238 --
## + attr: name (v/c), Type (e/c), weight (e/n), book (e/n),
## | source.family (e/c), target.family (e/c)
## + edges from 21f2988 (vertex names):
## [1] Arya-Stark--Benjen-Stark Arya-Stark--Bran-Stark
## [3] Arya-Stark--Catelyn-Stark Arya-Stark--Cersei-Lannister
## [5] Arya-Stark--Desmond Arya-Stark--Eddard-Stark
## [7] Arya-Stark--Ilyn-Payne Arya-Stark--Jeyne-Poole
## [9] Arya-Stark--Joffrey-Baratheon Arya-Stark--Jon-Snow
## [11] Arya-Stark--Jory-Cassel Arya-Stark--Meryn-Trant
## [13] Arya-Stark--Mordane Arya-Stark--Mycah
## + ... omitted several edges2.3.4 Visualization
- Plotting parameters: mapping important attributes to visual properties
- Find a good layout
?igraph.plotting2.3.4.1 Plotting parameters
](images/node.png)
Figure 2.8: Introduction from Kateto tutorial
](images/edge.png)
Figure 2.9: Introduction from Kateto tutorial
](images/other.png)
Figure 2.10: Introduction from Kateto tutorial
plot(got2, vertex.color="gold", vertex.size=3,
vertex.frame.color="gray", vertex.label.color="black",
vertex.label.cex=.5, vertex.label.dist=2, edge.curved=0.5,layout=layout_with_lgl)
2.3.4.1.1 To make the graph look nicer
- Node color: using family name
- Node size: degree
- Edge width: weight
## store the fullname
fullnames=V(got2)$name
fullnames[1:3]## [1] "Arya-Stark" "Benjen-Stark" "Bran-Stark"#get family name
familynames=fullnames%>%str_split("-",simplify = TRUE)%>%.[,2]
familynames[familynames==""]="None"
familynames[familynames=="(guard)"]="None"
# add vertices attributes
V(got2)$familyname=familynames
V(got2)$fullname=fullnames
V(got2)$name=nameshort # first nameSet colors and legend.
- pch: plotting symbols appearing in the legend
- pt.bg: background color for point
- cex: text size
- pt.cex: point size
- ncol: number of columns of the legend
- bty: “o”– rectangle box; “n” – no box
vcol=V(got2)$familyname
vcol[(vcol!="Stark")&(vcol!="Lannister")]="gray50"
vcol[vcol=="Stark"]="tomato"
vcol[vcol=="Lannister"]="gold"
V(got2)$color=vcol
V(got2)$size=igraph::degree(got2)%>%log()*4
E(got2)$width=E(got2)$weight%>%log()/2
plot(got2, vertex.label.color="black",
vertex.label.cex=.5, vertex.label.dist=2, edge.curved=0.5,layout=layout_with_kk)
legend("right", legend = c("Stark","Lannister","Other"), pch=21,
col=c("tomato","gold","gray50"), pt.bg=c("tomato","gold","gray50"), pt.cex=1, cex=.8, bty="n", ncol=1)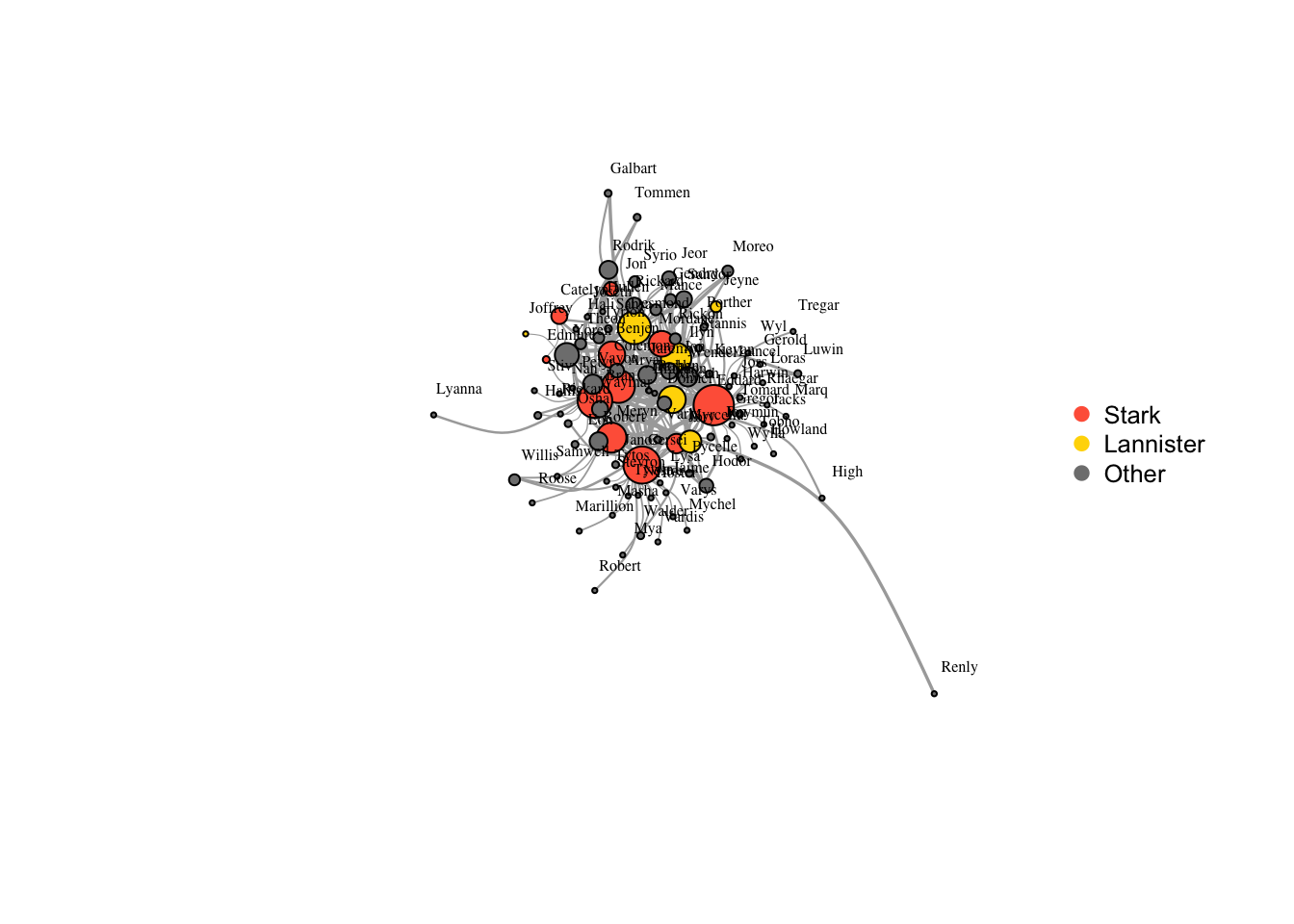
Plot only labels of the nodes
plot(got2, vertex.shape="none",vertex.label.color="black",
vertex.label.cex=.5, vertex.label.dist=2, edge.curved=0.5,layout=layout_with_kk)
2.3.4.2 Layouts
](images/layouts.png)
Figure 2.11: Layouts from Kateto tutorial
Force-directed layouts: suitable for general, small to medium sized graphs. (computational complexity; based on physical analogies)
- layout_with_fr: Fruchterman-Reingold is one of the most used force-directed layout algorithms. Force-directed layouts try to get a nice-looking graph where edges are similar in length and cross each other as little as possible. As a result, nodes are evenly distributed through the chart area, and the layout is intuitive in that nodes which share more connections are closer to each other.
- layout_with_kk: Another popular force-directed algorithm that produces nice results for connected graphs is Kamada Kawai.
- layout_with_graphopt: …
For large graphs:
- layout_with_lgl: The LGL algorithm is meant for large, connected graphs. Here you can also specify a root: a node that will be placed in the middle of the layout.
- layout_with_drl:
- layout_with_gfr:
- layout_with_dh:simulated annealing algorithm by Davidson and Harel
#layout_with_dh
plot(got2, vertex.label.color="black",
vertex.label.cex=.5,vertex.label.dist=0.2, edge.curved=0.5,layout=layout_with_dh)
legend("right", legend = c("Stark","Lannister","Other"), pch=21,
col=c("tomato","gold","gray50"), pt.bg=c("tomato","gold","gray50"), pt.cex=1, cex=.8, bty="n", ncol=1)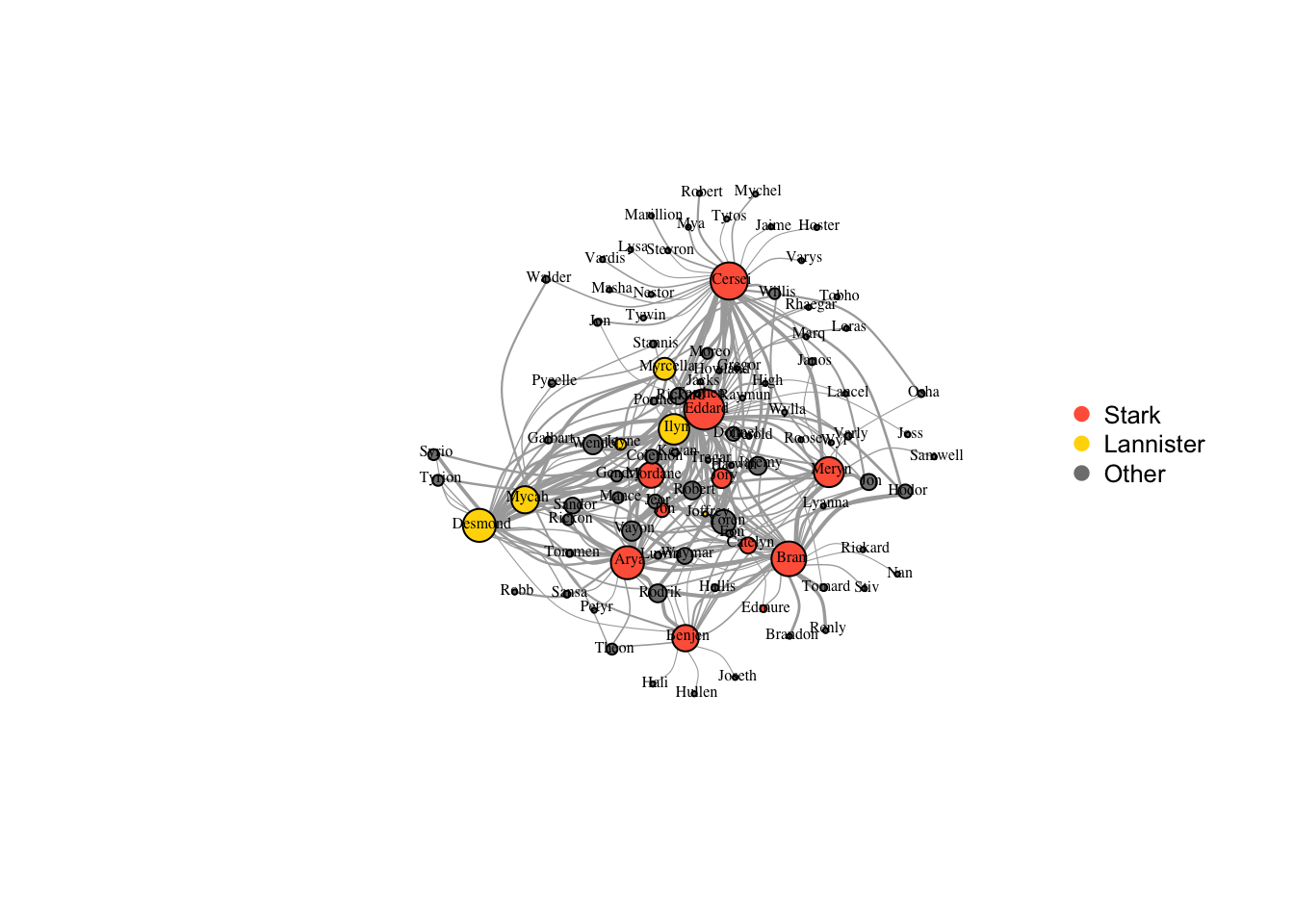
Selecting a layout automatically
- connected and vcount<=100: kk
- vcount<=1000:fr
- else: drl
plot(got2, vertex.label.color="black",
vertex.label.cex=.5,vertex.label.dist=0.2, edge.curved=0.5,layout=layout.auto(got2))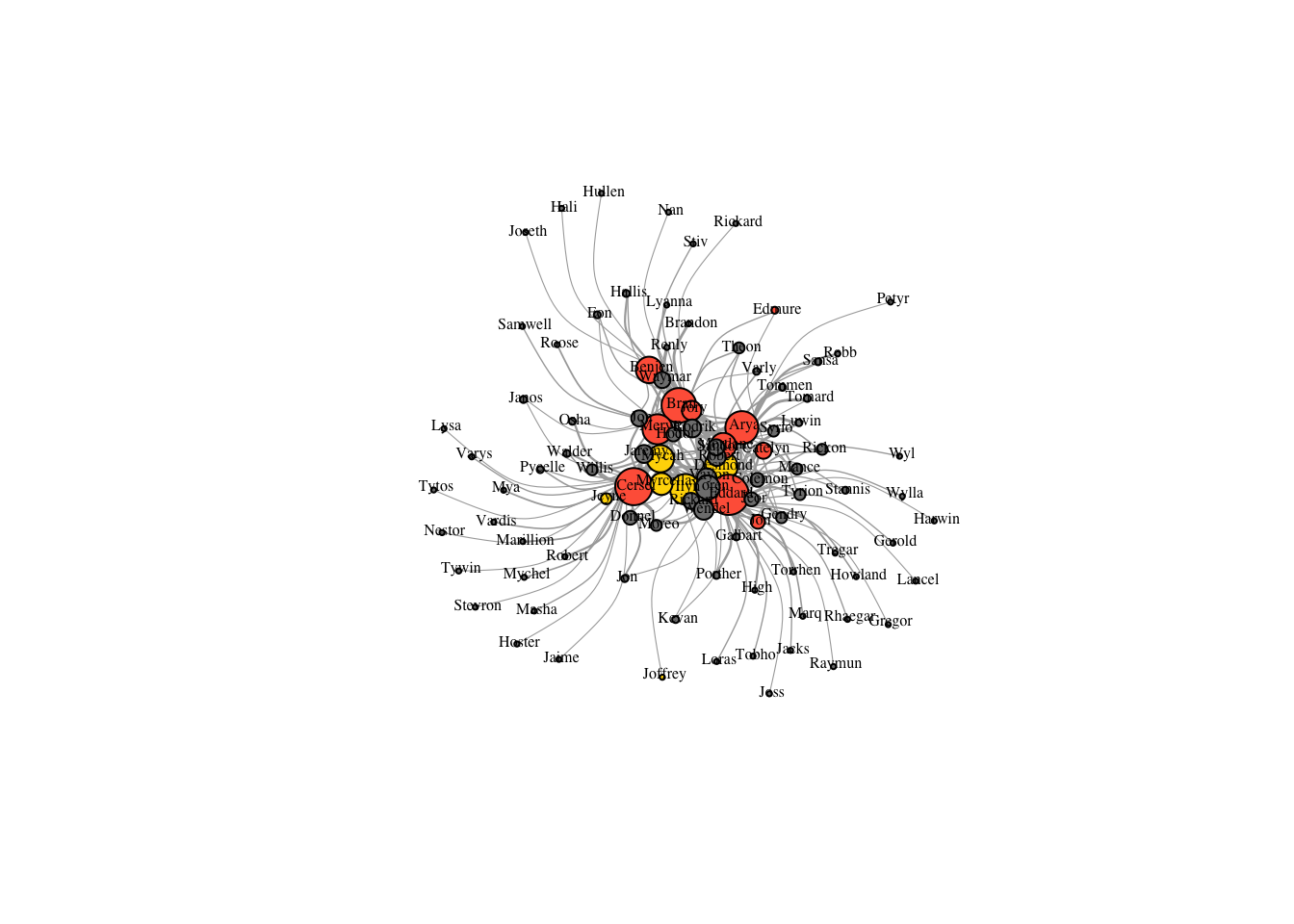
Without label and color the edge.
set.seed(2)
plot(got2, vertex.shape="none",vertex.label.color="black",
vertex.label.cex=.5,vertex.label.dist=0.2, edge.curved=0.5,layout=layout_with_dh)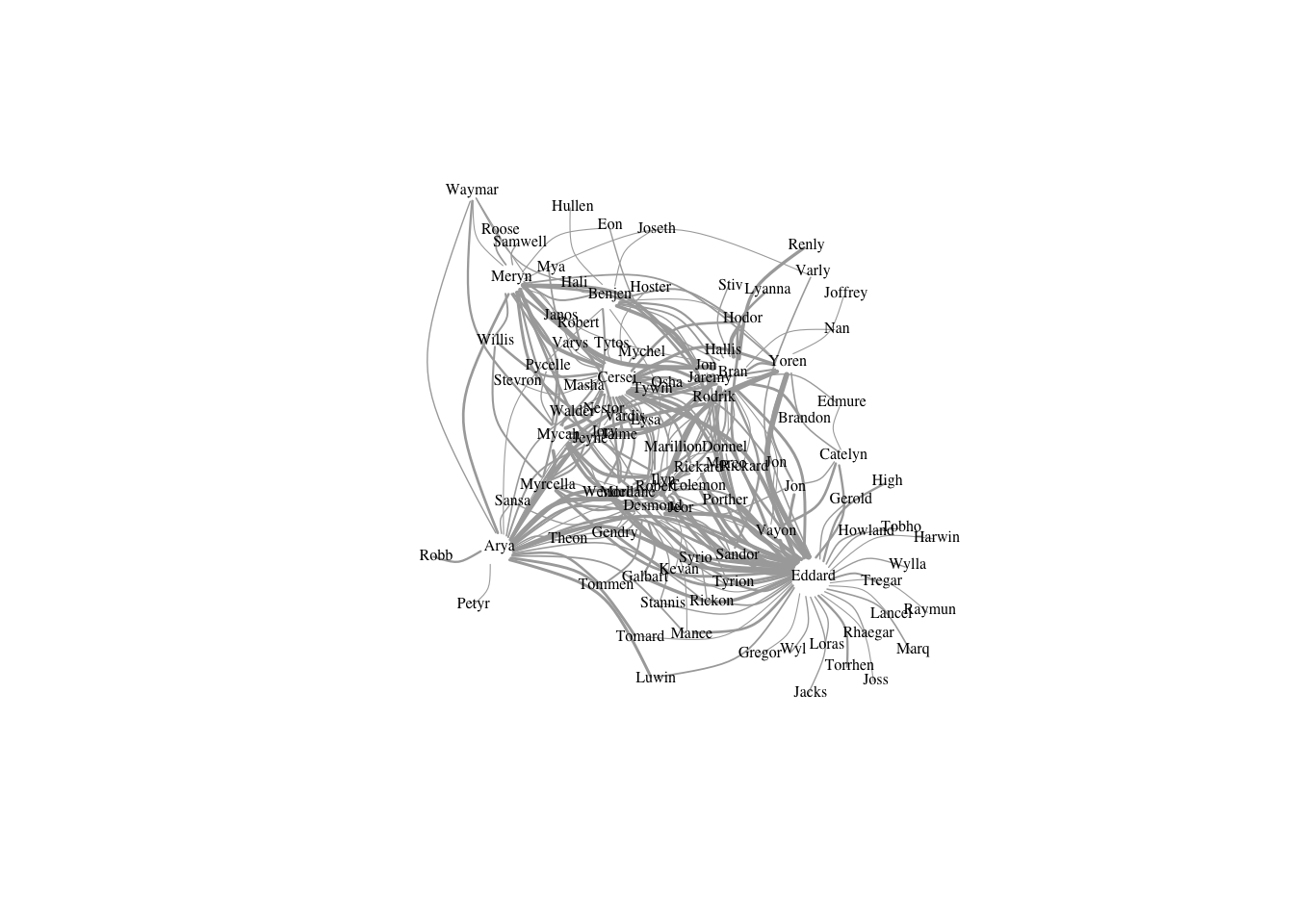
##color the edge
got2## IGRAPH 21f2988 UNW- 99 238 --
## + attr: name (v/c), familyname (v/c), fullname (v/c), color (v/c),
## | size (v/n), Type (e/c), weight (e/n), book (e/n), source.family
## | (e/c), target.family (e/c), width (e/n)
## + edges from 21f2988 (vertex names):
## [1] Arya--Benjen Arya--Bran Arya--Cersei Arya--Desmond Arya--Petyr
## [6] Arya--Eddard Arya--Rickon Arya--Robb Arya--Robert Arya--Rodrik
## [11] Arya--Sandor Arya--Sansa Arya--Syrio Arya--Tomard Arya--Tommen
## [16] Arya--Vayon Arya--Jory Arya--Meryn Arya--Yoren Arya--Jaremy
## [21] Arya--Jeor Arya--Mordane Arya--Luwin Arya--Mance Arya--Theon
## [26] Arya--Tyrion Arya--Waymar
## + ... omitted several edgesecol=rep("gray50",ecount(got2))
ecol[E(got2)$source.family=="Stark"]="tomato"
ecol[E(got2)$source.family=="Lannister"]="gold"
ecol[(ecol=="tomato")&(E(got2)$target.family=="Lannister")&(!is.na(E(got2)$target.family))]="orange"
ecol[(ecol=="gold")&(E(got2)$target.family=="Stark")&(!is.na(E(got2)$target.family))]="orange"
set.seed(2)
plot(got2, vertex.shape="none",vertex.label.color="black", edge.color=ecol,
vertex.label.cex=.5,vertex.label.dist=0.2, edge.curved=0.5,layout=layout_with_dh)
legend("right", legend = c("Stark","Lannister","Stark-Lannister","Other"),
col=c("tomato","gold","orange","gray50"), lty=rep(1,4), cex=.8, bty="n", ncol=1)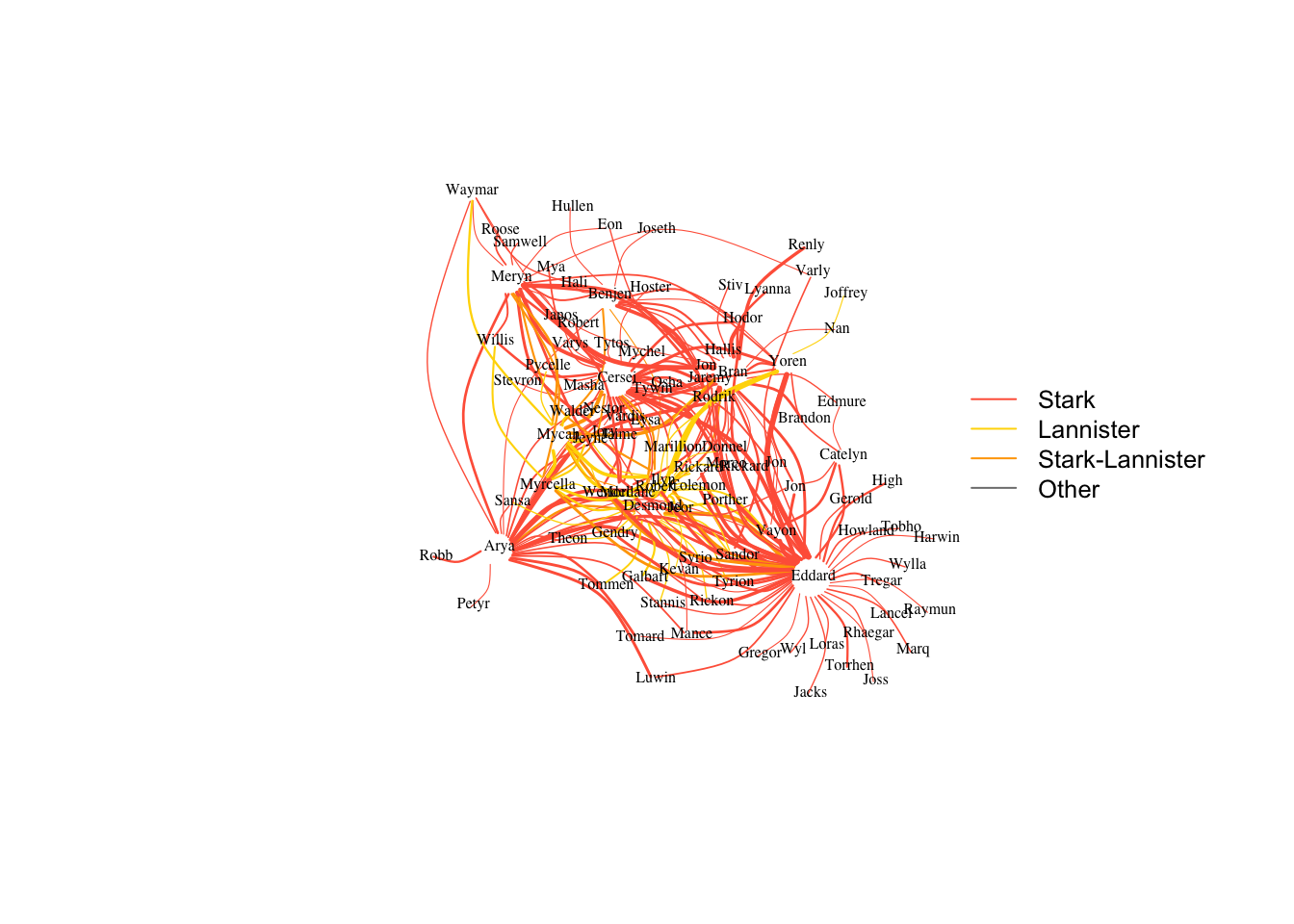
2.3.4.3 layout is not deterministic
Different runs will result in slightly different configurations. Saving the layout or set.seed allows us to get the exact same result multiple times, which can be helpful if you want to plot the time evolution of a graph, or different relationships – and want nodes to stay in the same place in multiple plots.
set.seed(1)
l=layout_with_dh(got2)
plot(got2, vertex.shape="none",vertex.label.color="black",
vertex.label.cex=.5,vertex.label.dist=0.2, edge.curved=0.5,layout=l)
rescale
norm_coordsrescale=F- can use
layout=l*2
l=layout_with_fr(got2)
l <- norm_coords(l, ymin=-1, ymax=1, xmin=-1, xmax=1) #default -- scaled
plot(got2, vertex.shape="none",vertex.label.color="black",
vertex.label.cex=.5,vertex.label.dist=0.2, edge.curved=0.5,layout=l,rescale=F)
Will introduce interactive r packages next time.
par(mfrow=c(2,2), mar=c(0,0,0,0))
plot(got2, vertex.shape="none",vertex.label.color="black",
vertex.label.cex=.5,vertex.label.dist=0.2, edge.curved=0.5,layout=l*0.5,rescale=F)
plot(got2, vertex.shape="none",vertex.label.color="black",
vertex.label.cex=.5,vertex.label.dist=0.2, edge.curved=0.5,layout=l*0.8,rescale=F)
plot(got2, vertex.shape="none",vertex.label.color="black",
vertex.label.cex=.5,vertex.label.dist=0.2, edge.curved=0.5,layout=l*1,rescale=F)
plot(got2, vertex.shape="none",vertex.label.color="black",
vertex.label.cex=.5,vertex.label.dist=0.2, edge.curved=0.5,layout=l*2,rescale=F)
#dev.off()2.3.5 Network and node descriptions
- Density:
edge_density - Degree:
degree - centrality and centralization:
centr_degree
closeness,centr_cloeigen_centrality,centr_eigenbetweenness,edge_betweenness,centr_betw
- reciprocity,transitivity,diameter,…
2.3.5.1 Density
The proportion of present edges from all possible ties.
edge_density(got2, loops=F)## [1] 0.04906205ecount(got2)/(vcount(got2)*(vcount(got2)-1))*2 #for an undirected network## [1] 0.049062052.3.5.2 Node degrees
‘degree’ has a mode of ‘in’ for in-degree, ‘out’ for out-degree, and ‘all’ or ‘total’ for total degree.
Notice the graph is undirected. So there is no difference under different parameter setting.
deg <- igraph::degree(got2, mode="all")
hist(deg, breaks=1:vcount(got2)-1, main="Histogram of node degree")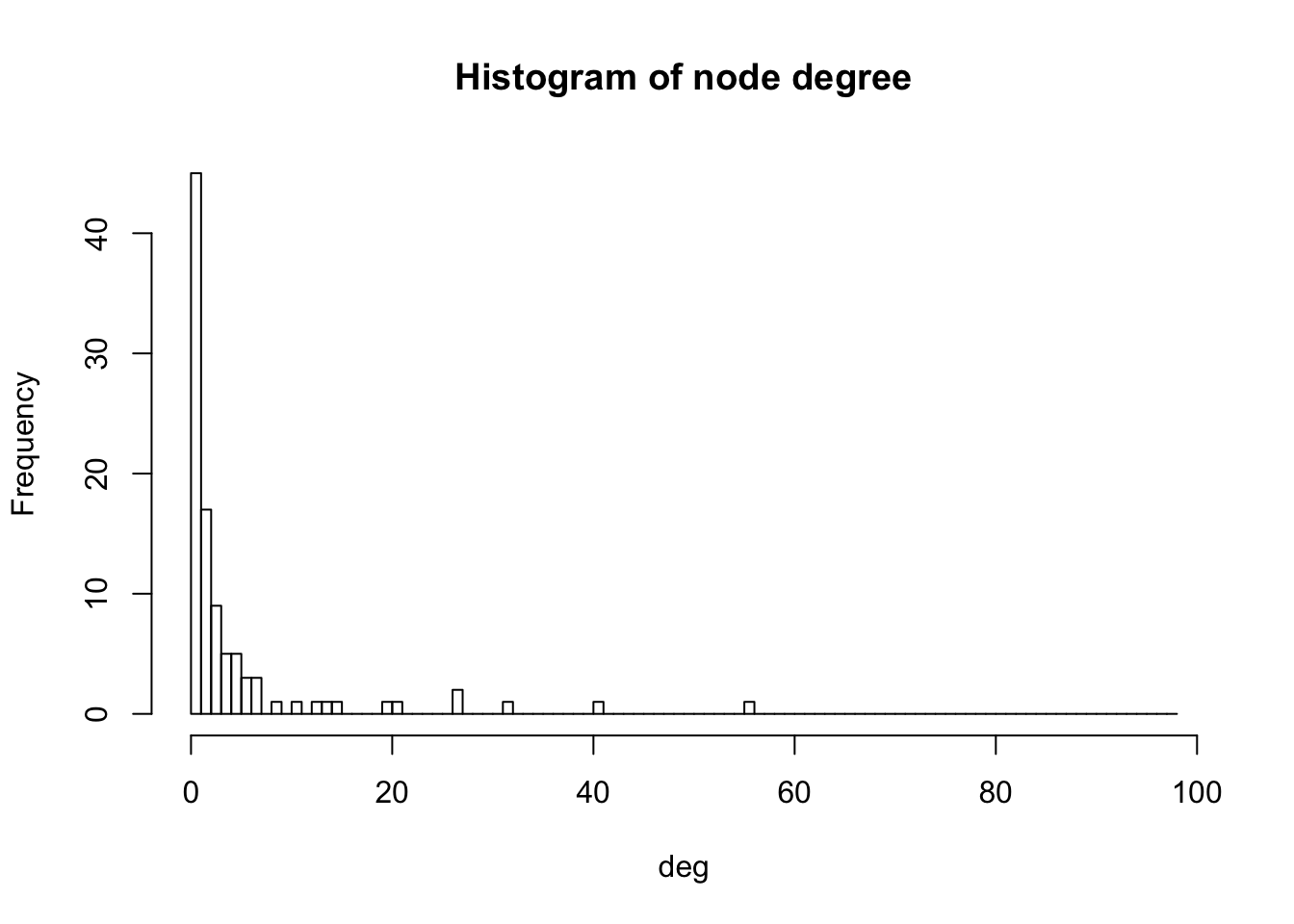
deg.dist <- degree_distribution(got2, cumulative=T, mode="all")
plot( x=0:max(deg), y=1-deg.dist, pch=19, cex=1.2, col="orange",
xlab="Degree", ylab="Cumulative Frequency")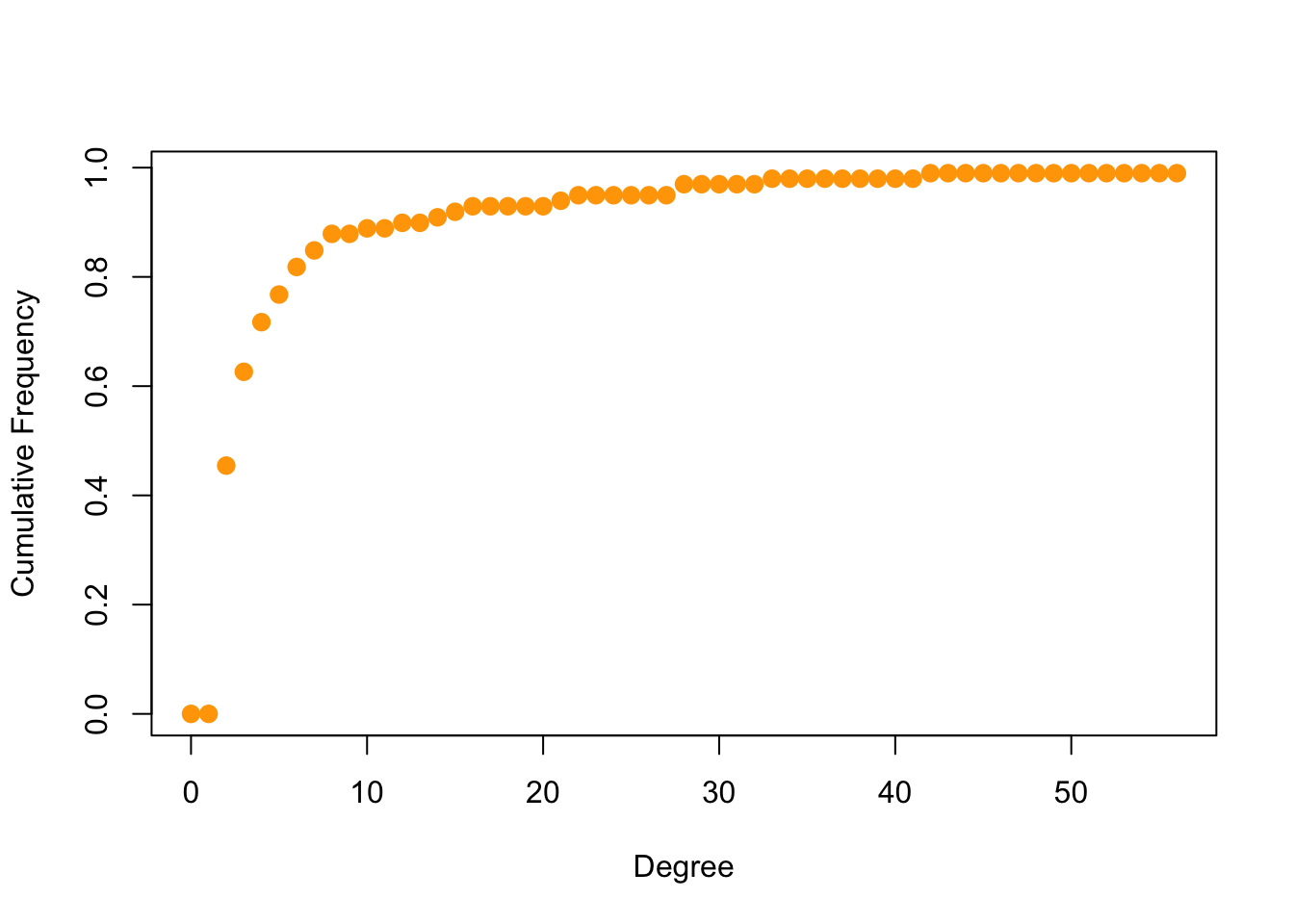
2.3.5.3 centrality and centralization
Who is the most important character?
- Degree
- Closeness
- Eigenvector
- Betweeness
Degree (number of ties).
Normalization should be the max degree the network can get
igraph::degree(got2, mode="in",loops = F)%>%sort(decreasing = TRUE)%>%.[1:5]## Eddard Cersei Bran Arya Desmond
## 56 41 32 27 27#Notice this is undirected network, the choice of mode does not matter
centr_degree(got2, mode="in", normalized=T,loops = F)$res%>%sort(decreasing = TRUE)%>%.[1:5]## [1] 56 41 32 27 27centr_degree(got2, mode="all", normalized=T,loops = F)$res%>%sort(decreasing = TRUE)%>%.[1:5]## [1] 56 41 32 27 27#Pay attention to whether allowing self-loop or not
# Normalization may differ due to the setting
centr_degree(got2, mode="all", normalized=T,loops = F)$theoretical_max## [1] 9506centr_degree(got2, mode="in", normalized=T,loops = F)$theoretical_max## [1] 9506centr_degree(got2, mode="in", normalized=T,loops = T)$theoretical_max## [1] 9702Closeness (centrality based on distance to others in the graph) Inverse of the node’s average geodesic distance to others in the network
#whether to include weight or not
#If a graph has edge attribute weight, the weight will be automatically took into consideration
igraph::closeness(got2, mode="all", weights=NA) %>%sort(decreasing = TRUE)%>%.[1:5]## Eddard Cersei Bran Arya Desmond
## 0.006993007 0.006329114 0.006097561 0.005882353 0.005847953igraph::closeness(got2, mode="all")%>%sort(decreasing = TRUE)%>%.[1:5]## Eddard Cersei Donnel Bran Arya
## 0.0010193680 0.0010111223 0.0010070493 0.0009990010 0.0009852217centr_clo(got2, mode="all", normalized=T)$res %>%sort(decreasing = TRUE)%>%.[1:5]## [1] 0.6853147 0.6202532 0.5975610 0.5764706 0.5730994Eigenvector (centrality proportional to the sum of connection centralities) Values of the first eigenvector of the graph adjacency matrix
eigen_centrality(got2, directed=F, weights=NA)$vector%>%sort(decreasing = TRUE)%>%.[1:5]## Eddard Cersei Bran Desmond Arya
## 1.0000000 0.8163499 0.7410532 0.7276696 0.6740883eigen_centrality(got2, directed=F)$vector%>%sort(decreasing = TRUE)%>%.[1:5]## Eddard Yoren Desmond Cersei Vayon
## 1.0000000 0.8538947 0.4281666 0.3352669 0.2441671centr_eigen(got2, directed=F, normalized=T) $vector%>%sort(decreasing = TRUE)%>%.[1:5]## [1] 1.0000000 0.8163499 0.7410532 0.7276696 0.6740883Betweenness (centrality based on a broker position connecting others) (Number of geodesics that pass through the node or the edge)
igraph::betweenness(got2, directed=F, weights=NA)%>%sort(decreasing = TRUE)%>%.[1:5]## Eddard Cersei Bran Arya Meryn
## 2155.2656 1554.1678 915.6561 510.5637 366.8074igraph::betweenness(got2, directed=F)%>%sort(decreasing = TRUE)%>%.[1:5]## Eddard Cersei Bran Benjen Arya
## 1835.5000 1483.2500 1024.8571 694.4762 689.5833edge_betweenness(got2, directed=F, weights=NA)%>%sort(decreasing = TRUE)%>%.[1:5]## [1] 426.4643 271.6982 198.3379 150.0371 133.8635centr_betw(got2, directed=F, normalized=T)$res%>%sort(decreasing = TRUE)%>%.[1:5]## [1] 2155.2656 1554.1678 915.6561 510.5637 366.80742.3.5.4 Other properties
- transitivity
- reciprocity
- clustering coefficient
- …
2.5 More about igraph
- Epidemics on networks: compartmental models on netwoks
- Spectral embeddings: community detection
- Change-point detection in temporal graphs
- CLustering multiple graphs
- Cliques and graphlets
- Graphons
- Graph matching