Chapter 3 ERGM (statnet Package)
3.1 Introduction
3.1.1 Outline
Here we only focus on the R package statnet to fit ERGM.
summarynetwork statisticsergmmodel fitting and interpretation:simulatenetwork simulations based on specified model.gof,mcmc.diagnostics: Goodness of fit and MCMC diagnostics
3.1.2 statnet
The analytic framework is based on Exponential family Random Graph Models (ergm).
statnet provides a comprehensive framework for ergm-based network modeling, including tools for:
- model estimation
- model-based network simulation
- network visualization
- model evaluation
Powered by Markov chain Monte Carlo (MCMC) algorithm.
3.1.3 References
- Official website (handbook): http://statnet.org/
- Tutorial: https://statnet.org/trac/raw-attachment/wiki/Sunbelt2016/ergm_tutorial.html
- Explore the official websites to find more info
3.2 Preparation
#install.packages("statnet")
library(statnet)## Installed ReposVer Built
## network "1.13.0.1" "1.15" "3.5.0"
## networkDynamic "0.9.0" "0.10.0" "3.5.0"library(dplyr)3.3 summary the network statistics
3.3.1 Dataset
florentine: Florentine Family Marriage and Business Ties Data
data(package="ergm") #see all available dataset in the package
data(florentine) # load flomarriage and flobusiness data
flomarriage # see the details of the network## Network attributes:
## vertices = 16
## directed = FALSE
## hyper = FALSE
## loops = FALSE
## multiple = FALSE
## bipartite = FALSE
## total edges= 20
## missing edges= 0
## non-missing edges= 20
##
## Vertex attribute names:
## priorates totalties vertex.names wealth
##
## No edge attributesclass(flomarriage)## [1] "network"summary(flomarriage)## Network attributes:
## vertices = 16
## directed = FALSE
## hyper = FALSE
## loops = FALSE
## multiple = FALSE
## bipartite = FALSE
## total edges = 20
## missing edges = 0
## non-missing edges = 20
## density = 0.1666667
##
## Vertex attributes:
##
## priorates:
## numeric valued attribute
## attribute summary:
## Min. 1st Qu. Median Mean 3rd Qu. Max.
## 0.00 0.00 21.50 25.94 44.75 74.00
##
## totalties:
## numeric valued attribute
## attribute summary:
## Min. 1st Qu. Median Mean 3rd Qu. Max.
## 1.00 4.75 9.00 13.88 15.00 54.00
## vertex.names:
## character valued attribute
## 16 valid vertex names
##
## wealth:
## numeric valued attribute
## attribute summary:
## Min. 1st Qu. Median Mean 3rd Qu. Max.
## 3.00 17.50 39.00 42.56 48.25 146.00
##
## No edge attributes
##
## Network edgelist matrix:
## [,1] [,2]
## [1,] 1 9
## [2,] 2 6
## [3,] 2 7
## [4,] 2 9
## [5,] 3 5
## [6,] 3 9
## [7,] 4 7
## [8,] 4 11
## [9,] 4 15
## [10,] 5 11
## [11,] 5 15
## [12,] 7 8
## [13,] 7 16
## [14,] 9 13
## [15,] 9 14
## [16,] 9 16
## [17,] 10 14
## [18,] 11 15
## [19,] 13 15
## [20,] 13 16set.seed(1)
plot(flomarriage)
3.3.2 summary
summary(network-object~ergm-terms): provide the statistics of network
summary(flomarriage~edges+triangle+kstar(1:3)+degree(0:5))## edges triangle kstar1 kstar2 kstar3 degree0 degree1 degree2
## 20 3 40 47 34 1 4 2
## degree3 degree4 degree5
## 6 2 03.3.3 directed networks
data(samplk)
samplk3## Network attributes:
## vertices = 18
## directed = TRUE
## hyper = FALSE
## loops = FALSE
## multiple = FALSE
## bipartite = FALSE
## total edges= 56
## missing edges= 0
## non-missing edges= 56
##
## Vertex attribute names:
## cloisterville group vertex.names
##
## No edge attributesset.seed(1)
plot(samplk3)
idegree and odegree.
summary(samplk3~idegree(1:3)+odegree(c(2,4))) #indegree and outdegree## idegree1 idegree2 idegree3 odegree2 odegree4
## 1 7 2 0 23.4 ergm fit ergm model
3.4.1 ERGM
The general form: \(P(g=G)=\frac{exp(\theta'T(G))}{c(\theta)}\)
log-odds of a single edge between node \(i\) and \(j\): \(logit(Y)=\theta'T(G)\)
Similar as regression:
glm(y~x) – ergm(graph~terms) where terms are \(T(\cdot)\)
3.4.2 ergm function.
- Input: formula [netowrk object (adjacency matrix)] ~ [ergm-terms]
#Input
?ergm
class(flomarriage)## [1] "network"ergm(flomarriage~edges)%>%summary()## Starting maximum pseudolikelihood estimation (MPLE):## Evaluating the predictor and response matrix.## Maximizing the pseudolikelihood.## Finished MPLE.## Stopping at the initial estimate.## Evaluating log-likelihood at the estimate.##
## ==========================
## Summary of model fit
## ==========================
##
## Formula: flomarriage ~ edges
##
## Iterations: 5 out of 20
##
## Monte Carlo MLE Results:
## Estimate Std. Error MCMC % z value Pr(>|z|)
## edges -1.6094 0.2449 0 -6.571 <1e-04 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Null Deviance: 166.4 on 120 degrees of freedom
## Residual Deviance: 108.1 on 119 degrees of freedom
##
## AIC: 110.1 BIC: 112.9 (Smaller is better.)mat=as.sociomatrix(flomarriage)
ergm(mat~edges)%>%summary()## Starting maximum pseudolikelihood estimation (MPLE):
## Evaluating the predictor and response matrix.
## Maximizing the pseudolikelihood.
## Finished MPLE.
## Stopping at the initial estimate.
## Evaluating log-likelihood at the estimate.##
## ==========================
## Summary of model fit
## ==========================
##
## Formula: mat ~ edges
##
## Iterations: 5 out of 20
##
## Monte Carlo MLE Results:
## Estimate Std. Error MCMC % z value Pr(>|z|)
## edges -1.6094 0.1732 0 -9.292 <1e-04 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Null Deviance: 332.7 on 240 degrees of freedom
## Residual Deviance: 216.3 on 239 degrees of freedom
##
## AIC: 218.3 BIC: 221.8 (Smaller is better.)Output: ergm object
Method on ergm object:
- class
- names
- summary
- coef
- coefficients
- vcov
flomodel.01=ergm(flomarriage~edges)## Starting maximum pseudolikelihood estimation (MPLE):## Evaluating the predictor and response matrix.## Maximizing the pseudolikelihood.## Finished MPLE.## Stopping at the initial estimate.## Evaluating log-likelihood at the estimate.summary(flomodel.01)##
## ==========================
## Summary of model fit
## ==========================
##
## Formula: flomarriage ~ edges
##
## Iterations: 5 out of 20
##
## Monte Carlo MLE Results:
## Estimate Std. Error MCMC % z value Pr(>|z|)
## edges -1.6094 0.2449 0 -6.571 <1e-04 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Null Deviance: 166.4 on 120 degrees of freedom
## Residual Deviance: 108.1 on 119 degrees of freedom
##
## AIC: 110.1 BIC: 112.9 (Smaller is better.)class(flomodel.01)## [1] "ergm"names(flomodel.01)## [1] "coef" "iterations" "MCMCtheta" "gradient"
## [5] "hessian" "covar" "failure" "est.cov"
## [9] "glm" "glm.null" "theta1" "offset"
## [13] "drop" "estimable" "network" "reference"
## [17] "newnetwork" "formula" "constrained" "constraints"
## [21] "target.stats" "etamap" "target.esteq" "estimate"
## [25] "control" "null.lik" "mle.lik"flomodel.01$coef## edges
## -1.609438coef(flomodel.01)## edges
## -1.609438coefficients(flomodel.01)## Warning: You appear to be calling coef.ergm() directly. coef.ergm() is a
## method, and will not be exported in a future version of 'ergm'. Use coef()
## instead, or getS3method() if absolutely necessary.## edges
## -1.609438vcov(flomodel.01) #variance-covariance matrix of the main parameters## edges
## edges 0.05999985vcov(flomodel.01)%>%sqrt()## edges
## edges 0.2449487vcov(ergm(flomarriage~edges+triangle))## Starting maximum pseudolikelihood estimation (MPLE):
## Evaluating the predictor and response matrix.
## Maximizing the pseudolikelihood.
## Finished MPLE.
## Starting Monte Carlo maximum likelihood estimation (MCMLE):
## Iteration 1 of at most 20:
## Optimizing with step length 1.
## The log-likelihood improved by 0.002922.
## Step length converged once. Increasing MCMC sample size.
## Iteration 2 of at most 20:
## Optimizing with step length 1.
## The log-likelihood improved by 0.0006953.
## Step length converged twice. Stopping.
## Finished MCMLE.
## Evaluating log-likelihood at the estimate. Using 20 bridges: 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 .
## This model was fit using MCMC. To examine model diagnostics and check for degeneracy, use the mcmc.diagnostics() function.## edges triangle
## edges 0.1242829 -0.1573260
## triangle -0.1573260 0.35390893.4.3 ergm-terms
See the full list of ergm-terms
help('ergm-terms')3.4.3.1 Erdos-Renyi model G(n,p) model: include edges term
Edge included in the graph with prob \(p\): \(Pr(A=a|p)=\prod p^{a_{ij}}(1-p)^{1-a_{ij}}\)
Rewrite Erdos-Renyi model in ERGM form: \(T(G)=\#edges\), \(\theta=log \frac{p}{1-p}\)
3.4.3.2 Interpretation of the parameter
Interpretation (similar when you interpret glm):
edges: If the number of edges increase 1, the log-odds of any edge existing is -1.6094.\(p<\alpha\): parameter does not equal to 0 significantly,
Null Deviance: The null deviance in the ergm output appears to be based on an Erdos-Renyi random graph with p = 0.5.
Residual Deviance: 2 times difference of loglik (saturated model - our model) (smaller is better)
AIC, BIC: -loglik+penalty(#parameter)
flomodel.01=ergm(flomarriage~edges) #similar as regression lm(y~x)## Starting maximum pseudolikelihood estimation (MPLE):## Evaluating the predictor and response matrix.## Maximizing the pseudolikelihood.## Finished MPLE.## Stopping at the initial estimate.## Evaluating log-likelihood at the estimate.summary(flomodel.01) ##
## ==========================
## Summary of model fit
## ==========================
##
## Formula: flomarriage ~ edges
##
## Iterations: 5 out of 20
##
## Monte Carlo MLE Results:
## Estimate Std. Error MCMC % z value Pr(>|z|)
## edges -1.6094 0.2449 0 -6.571 <1e-04 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Null Deviance: 166.4 on 120 degrees of freedom
## Residual Deviance: 108.1 on 119 degrees of freedom
##
## AIC: 110.1 BIC: 112.9 (Smaller is better.)flomodel.01$coef/(1+exp(flomodel.01$coef)) # prob of an edge exists## edges
## -1.341198-2*(flomodel.01$mle.lik)+2 #AIC: -2loglik+2k## 'log Lik.' 110.1347 (df=1)-2*(flomodel.01$mle.lik)+log(16*15/2) #BIC:-2loglik+klog(n)## 'log Lik.' 112.9222 (df=1)pchisq(108.1 , df=119, lower.tail=FALSE) # H0: our model fits well. The smaller the better. Accept H0.## [1] 0.75357283.4.3.3 Include triangle terms
Include number of triangles as a measure of clustering.
flomodel.02=ergm(flomarriage~edges+triangle)## Starting maximum pseudolikelihood estimation (MPLE):## Evaluating the predictor and response matrix.## Maximizing the pseudolikelihood.## Finished MPLE.## Starting Monte Carlo maximum likelihood estimation (MCMLE):## Iteration 1 of at most 20:## Optimizing with step length 1.## The log-likelihood improved by 0.00888.## Step length converged once. Increasing MCMC sample size.## Iteration 2 of at most 20:## Optimizing with step length 1.## The log-likelihood improved by 0.000148.## Step length converged twice. Stopping.## Finished MCMLE.## Evaluating log-likelihood at the estimate. Using 20 bridges: 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 .
## This model was fit using MCMC. To examine model diagnostics and check for degeneracy, use the mcmc.diagnostics() function.summary(flomodel.02)##
## ==========================
## Summary of model fit
## ==========================
##
## Formula: flomarriage ~ edges + triangle
##
## Iterations: 2 out of 20
##
## Monte Carlo MLE Results:
## Estimate Std. Error MCMC % z value Pr(>|z|)
## edges -1.6737 0.3578 0 -4.677 <1e-04 ***
## triangle 0.1490 0.6029 0 0.247 0.805
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Null Deviance: 166.4 on 120 degrees of freedom
## Residual Deviance: 108.1 on 118 degrees of freedom
##
## AIC: 112.1 BIC: 117.6 (Smaller is better.)Coefficients: -1.6744\(\times\)#of edges+0.1497\(\times\)#of triangles
triangleis not significantAIC, BIC: larger than that of ER model
3.4.3.4 Include nodal covariates: nodecov
Using nodecov to include continuous nodal covariates
flomarriage## Network attributes:
## vertices = 16
## directed = FALSE
## hyper = FALSE
## loops = FALSE
## multiple = FALSE
## bipartite = FALSE
## total edges= 20
## missing edges= 0
## non-missing edges= 20
##
## Vertex attribute names:
## priorates totalties vertex.names wealth
##
## No edge attributeswealth=flomarriage %v% 'wealth' # %v% get the vertex attributes
set.seed(1)
plot(flomarriage, vertex.cex=wealth/25, main="Florentine marriage by wealth", cex.main=0.8)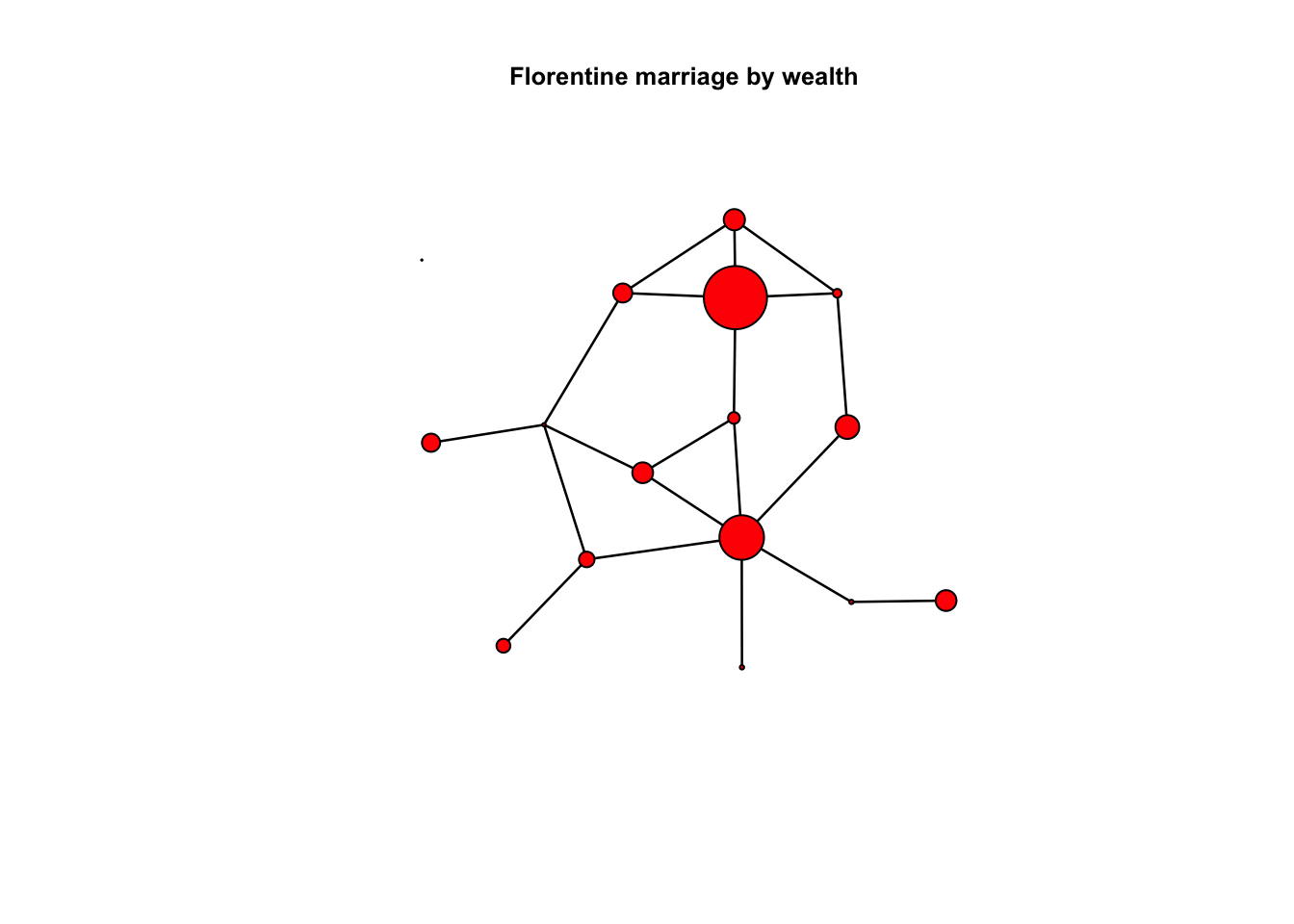
flomodel.03=ergm(flomarriage~edges+nodecov('wealth'))## Starting maximum pseudolikelihood estimation (MPLE):## Evaluating the predictor and response matrix.## Maximizing the pseudolikelihood.## Finished MPLE.## Stopping at the initial estimate.## Evaluating log-likelihood at the estimate.summary(flomodel.03)##
## ==========================
## Summary of model fit
## ==========================
##
## Formula: flomarriage ~ edges + nodecov("wealth")
##
## Iterations: 4 out of 20
##
## Monte Carlo MLE Results:
## Estimate Std. Error MCMC % z value Pr(>|z|)
## edges -2.594929 0.536056 0 -4.841 <1e-04 ***
## nodecov.wealth 0.010546 0.004674 0 2.256 0.0241 *
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Null Deviance: 166.4 on 120 degrees of freedom
## Residual Deviance: 103.1 on 118 degrees of freedom
##
## AIC: 107.1 BIC: 112.7 (Smaller is better.)Interpretation: -2.594929 # of edges + 0.010546 wealth of node i + 0.010546 wealth of node j
3.4.3.5 Include transformation of continuous nodal covariates
flomodel.04=ergm(flomarriage~edges+absdiff("wealth"))## Starting maximum pseudolikelihood estimation (MPLE):## Evaluating the predictor and response matrix.## Maximizing the pseudolikelihood.## Finished MPLE.## Stopping at the initial estimate.## Evaluating log-likelihood at the estimate.summary(flomodel.04)##
## ==========================
## Summary of model fit
## ==========================
##
## Formula: flomarriage ~ edges + absdiff("wealth")
##
## Iterations: 4 out of 20
##
## Monte Carlo MLE Results:
## Estimate Std. Error MCMC % z value Pr(>|z|)
## edges -2.302042 0.401906 0 -5.728 <1e-04 ***
## absdiff.wealth 0.015519 0.006157 0 2.521 0.0117 *
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Null Deviance: 166.4 on 120 degrees of freedom
## Residual Deviance: 102.0 on 118 degrees of freedom
##
## AIC: 106 BIC: 111.5 (Smaller is better.)3.4.3.6 Include transformation of continuous nodal covariates
flomodel.05=ergm(flomarriage~edges+nodecov('wealth')+nodecov('wealth',transform=function(x) x^2))## Starting maximum pseudolikelihood estimation (MPLE):## Evaluating the predictor and response matrix.## Maximizing the pseudolikelihood.## Finished MPLE.## Stopping at the initial estimate.## Evaluating log-likelihood at the estimate.summary(flomodel.05)##
## ==========================
## Summary of model fit
## ==========================
##
## Formula: flomarriage ~ edges + nodecov("wealth") + nodecov("wealth", transform = function(x) x^2)
##
## Iterations: 4 out of 20
##
## Monte Carlo MLE Results:
## Estimate Std. Error MCMC % z value Pr(>|z|)
## edges -2.904e+00 9.313e-01 0 -3.119 0.00182 **
## nodecov.wealth 1.730e-02 1.695e-02 0 1.021 0.30737
## nodecov.wealth.1 -4.459e-05 1.073e-04 0 -0.416 0.67768
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Null Deviance: 166.4 on 120 degrees of freedom
## Residual Deviance: 102.9 on 117 degrees of freedom
##
## AIC: 108.9 BIC: 117.3 (Smaller is better.)3.4.3.7 Include other possible ergm-terms
flomodel.06=ergm(flomarriage~kstar(1:2) + absdiff("wealth"))## Starting maximum pseudolikelihood estimation (MPLE):## Evaluating the predictor and response matrix.## Maximizing the pseudolikelihood.## Finished MPLE.## Starting Monte Carlo maximum likelihood estimation (MCMLE):## Iteration 1 of at most 20:## Optimizing with step length 1.## The log-likelihood improved by 0.01795.## Step length converged once. Increasing MCMC sample size.## Iteration 2 of at most 20:## Optimizing with step length 1.## The log-likelihood improved by 0.001918.## Step length converged twice. Stopping.## Finished MCMLE.## Evaluating log-likelihood at the estimate. Using 20 bridges: 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 .
## This model was fit using MCMC. To examine model diagnostics and check for degeneracy, use the mcmc.diagnostics() function.summary(flomodel.06)##
## ==========================
## Summary of model fit
## ==========================
##
## Formula: flomarriage ~ kstar(1:2) + absdiff("wealth")
##
## Iterations: 2 out of 20
##
## Monte Carlo MLE Results:
## Estimate Std. Error MCMC % z value Pr(>|z|)
## kstar1 -0.857799 0.455348 0 -1.884 0.0596 .
## kstar2 -0.162150 0.224192 0 -0.723 0.4695
## absdiff.wealth 0.019244 0.008719 0 2.207 0.0273 *
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Null Deviance: 166.4 on 120 degrees of freedom
## Residual Deviance: 101.3 on 117 degrees of freedom
##
## AIC: 107.3 BIC: 115.7 (Smaller is better.)summary(ergm(flomarriage~kstar(c(1,3)) + absdiff("wealth")))## Starting maximum pseudolikelihood estimation (MPLE):
## Evaluating the predictor and response matrix.
## Maximizing the pseudolikelihood.
## Finished MPLE.
## Starting Monte Carlo maximum likelihood estimation (MCMLE):
## Iteration 1 of at most 20:
## Optimizing with step length 1.
## The log-likelihood improved by 0.01691.
## Step length converged once. Increasing MCMC sample size.
## Iteration 2 of at most 20:
## Optimizing with step length 1.
## The log-likelihood improved by 0.001825.
## Step length converged twice. Stopping.
## Finished MCMLE.
## Evaluating log-likelihood at the estimate. Using 20 bridges: 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 .
## This model was fit using MCMC. To examine model diagnostics and check for degeneracy, use the mcmc.diagnostics() function.##
## ==========================
## Summary of model fit
## ==========================
##
## Formula: flomarriage ~ kstar(c(1, 3)) + absdiff("wealth")
##
## Iterations: 2 out of 20
##
## Monte Carlo MLE Results:
## Estimate Std. Error MCMC % z value Pr(>|z|)
## kstar1 -1.054277 0.261968 0 -4.024 <1e-04 ***
## kstar3 -0.086239 0.111865 0 -0.771 0.4408
## absdiff.wealth 0.020937 0.009522 0 2.199 0.0279 *
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Null Deviance: 166.4 on 120 degrees of freedom
## Residual Deviance: 101.0 on 117 degrees of freedom
##
## AIC: 107 BIC: 115.4 (Smaller is better.)3.4.3.8 Include categorical nodal covariates: nodefactor
data("faux.mesa.high")
mesa=faux.mesa.high
mesa # grade, race, sex are discrete vertex attribute## Network attributes:
## vertices = 205
## directed = FALSE
## hyper = FALSE
## loops = FALSE
## multiple = FALSE
## bipartite = FALSE
## total edges= 203
## missing edges= 0
## non-missing edges= 203
##
## Vertex attribute names:
## Grade Race Sex
##
## No edge attributesplot(mesa, vertex.col='Grade')
legend('bottomleft',fill=7:12,legend=paste('Grade',7:12),cex=0.75)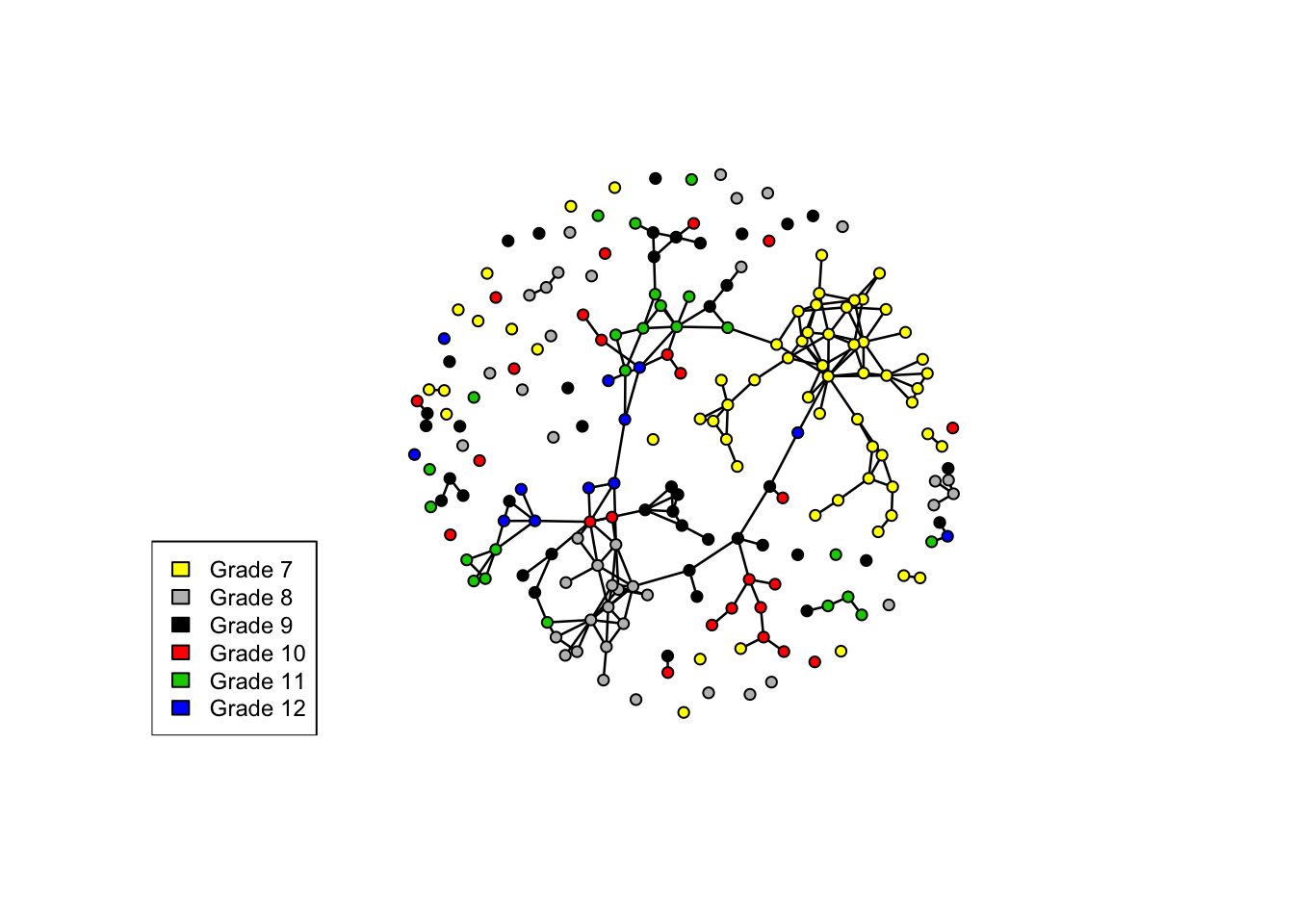
3.4.3.9 Include discrete nodal covariates: nodefactor
fauxmodel.01=ergm(mesa ~edges + nodecov('Grade'))## Starting maximum pseudolikelihood estimation (MPLE):## Evaluating the predictor and response matrix.## Maximizing the pseudolikelihood.## Finished MPLE.## Stopping at the initial estimate.## Evaluating log-likelihood at the estimate.summary(fauxmodel.01)##
## ==========================
## Summary of model fit
## ==========================
##
## Formula: mesa ~ edges + nodecov("Grade")
##
## Iterations: 7 out of 20
##
## Monte Carlo MLE Results:
## Estimate Std. Error MCMC % z value Pr(>|z|)
## edges -3.64578 0.56757 0 -6.424 <1e-04 ***
## nodecov.Grade -0.05650 0.03274 0 -1.726 0.0844 .
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Null Deviance: 28987 on 20910 degrees of freedom
## Residual Deviance: 2283 on 20908 degrees of freedom
##
## AIC: 2287 BIC: 2303 (Smaller is better.)fauxmodel.02=ergm(mesa ~edges + nodefactor('Grade'))## Starting maximum pseudolikelihood estimation (MPLE):
## Evaluating the predictor and response matrix.
## Maximizing the pseudolikelihood.
## Finished MPLE.
## Stopping at the initial estimate.
## Evaluating log-likelihood at the estimate.summary(fauxmodel.02)##
## ==========================
## Summary of model fit
## ==========================
##
## Formula: mesa ~ edges + nodefactor("Grade")
##
## Iterations: 6 out of 20
##
## Monte Carlo MLE Results:
## Estimate Std. Error MCMC % z value Pr(>|z|)
## edges -4.17784 0.14696 0 -28.428 < 1e-04 ***
## nodefactor.Grade.8 -0.27924 0.14209 0 -1.965 0.04938 *
## nodefactor.Grade.9 -0.47364 0.14912 0 -3.176 0.00149 **
## nodefactor.Grade.10 -0.54653 0.18641 0 -2.932 0.00337 **
## nodefactor.Grade.11 -0.19281 0.16550 0 -1.165 0.24401
## nodefactor.Grade.12 -0.05704 0.20739 0 -0.275 0.78330
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Null Deviance: 28987 on 20910 degrees of freedom
## Residual Deviance: 2269 on 20904 degrees of freedom
##
## AIC: 2281 BIC: 2329 (Smaller is better.)3.4.3.10 Include other terms: eg. homophily
help('ergm-terms') # check all the possible ergm terms3.4.3.11 Include other terms: eg. homophily
nodematch: Uniform homophily and differential homophily
uniform homophily(diff=FALSE), adds one network statistic to the model, which counts the number of edges (i,j) for which attrname(i)==attrname(j).
differential homophily(diff=TRUE), p(#of unique values of the attrname attribute) network statistics are added to the model.
The kth such statistic counts the number of edges (i,j) for which attrname(i) == attrname(j) == value(k), where value(k) is the kth smallest unique value of the attrname attribute.
When multiple attribute names are given, the statistic counts only ties for which all of the attributes match.
?nodematch
fauxmodel.03 <- ergm(mesa ~edges + nodematch('Grade',diff=F) )## Starting maximum pseudolikelihood estimation (MPLE):## Evaluating the predictor and response matrix.## Maximizing the pseudolikelihood.## Finished MPLE.## Stopping at the initial estimate.## Evaluating log-likelihood at the estimate.summary(fauxmodel.03)##
## ==========================
## Summary of model fit
## ==========================
##
## Formula: mesa ~ edges + nodematch("Grade", diff = F)
##
## Iterations: 8 out of 20
##
## Monte Carlo MLE Results:
## Estimate Std. Error MCMC % z value Pr(>|z|)
## edges -6.0340 0.1583 0 -38.12 <1e-04 ***
## nodematch.Grade 2.8310 0.1773 0 15.96 <1e-04 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Null Deviance: 28987 on 20910 degrees of freedom
## Residual Deviance: 1940 on 20908 degrees of freedom
##
## AIC: 1944 BIC: 1959 (Smaller is better.)mixingmatrix(mesa, "Race") # reason for Inf## Note: Marginal totals can be misleading
## for undirected mixing matrices.
## Black Hisp NatAm Other White
## Black 0 8 13 0 5
## Hisp 8 53 41 1 22
## NatAm 13 41 46 0 10
## Other 0 1 0 0 0
## White 5 22 10 0 4fauxmodel.04 <- ergm(mesa ~edges + nodematch('Grade',diff=T) )## Starting maximum pseudolikelihood estimation (MPLE):
## Evaluating the predictor and response matrix.
## Maximizing the pseudolikelihood.
## Finished MPLE.
## Stopping at the initial estimate.
## Evaluating log-likelihood at the estimate.summary(fauxmodel.04)##
## ==========================
## Summary of model fit
## ==========================
##
## Formula: mesa ~ edges + nodematch("Grade", diff = T)
##
## Iterations: 8 out of 20
##
## Monte Carlo MLE Results:
## Estimate Std. Error MCMC % z value Pr(>|z|)
## edges -6.0340 0.1583 0 -38.117 <1e-04 ***
## nodematch.Grade.7 2.8471 0.1973 0 14.427 <1e-04 ***
## nodematch.Grade.8 2.9145 0.2381 0 12.240 <1e-04 ***
## nodematch.Grade.9 2.4385 0.2641 0 9.234 <1e-04 ***
## nodematch.Grade.10 2.5579 0.3736 0 6.846 <1e-04 ***
## nodematch.Grade.11 3.3104 0.2962 0 11.176 <1e-04 ***
## nodematch.Grade.12 3.7315 0.4565 0 8.174 <1e-04 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Null Deviance: 28987 on 20910 degrees of freedom
## Residual Deviance: 1928 on 20903 degrees of freedom
##
## AIC: 1942 BIC: 1998 (Smaller is better.)fauxmodel.04 <- ergm(mesa ~edges + nodematch('Grade',diff=T,keep=c(2,4)) )## Starting maximum pseudolikelihood estimation (MPLE):
## Evaluating the predictor and response matrix.
## Maximizing the pseudolikelihood.
## Finished MPLE.
## Stopping at the initial estimate.
## Evaluating log-likelihood at the estimate.summary(fauxmodel.04)##
## ==========================
## Summary of model fit
## ==========================
##
## Formula: mesa ~ edges + nodematch("Grade", diff = T, keep = c(2, 4))
##
## Iterations: 7 out of 20
##
## Monte Carlo MLE Results:
## Estimate Std. Error MCMC % z value Pr(>|z|)
## edges -4.80539 0.07913 0 -60.727 < 1e-04 ***
## nodematch.Grade.8 1.68584 0.19469 0 8.659 < 1e-04 ***
## nodematch.Grade.10 1.32930 0.34758 0 3.824 0.000131 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Null Deviance: 28987 on 20910 degrees of freedom
## Residual Deviance: 2225 on 20907 degrees of freedom
##
## AIC: 2231 BIC: 2255 (Smaller is better.)fauxmodel.05 <- ergm(mesa ~edges + nodematch('Grade',diff=T)+ nodematch('Race',diff=T) )## Observed statistic(s) nodematch.Race.Black and nodematch.Race.Other are at their smallest attainable values. Their coefficients will be fixed at -Inf.
## Starting maximum pseudolikelihood estimation (MPLE):
## Evaluating the predictor and response matrix.
## Maximizing the pseudolikelihood.
## Finished MPLE.
## Stopping at the initial estimate.
## Evaluating log-likelihood at the estimate.summary(fauxmodel.05)##
## ==========================
## Summary of model fit
## ==========================
##
## Formula: mesa ~ edges + nodematch("Grade", diff = T) + nodematch("Race",
## diff = T)
##
## Iterations: 8 out of 20
##
## Monte Carlo MLE Results:
## Estimate Std. Error MCMC % z value Pr(>|z|)
## edges -6.2328 0.1742 0 -35.785 <1e-04 ***
## nodematch.Grade.7 2.8740 0.1981 0 14.509 <1e-04 ***
## nodematch.Grade.8 2.8788 0.2391 0 12.038 <1e-04 ***
## nodematch.Grade.9 2.4642 0.2647 0 9.310 <1e-04 ***
## nodematch.Grade.10 2.5692 0.3770 0 6.815 <1e-04 ***
## nodematch.Grade.11 3.2921 0.2978 0 11.057 <1e-04 ***
## nodematch.Grade.12 3.8376 0.4592 0 8.357 <1e-04 ***
## nodematch.Race.Black -Inf 0.0000 0 -Inf <1e-04 ***
## nodematch.Race.Hisp 0.0679 0.1737 0 0.391 0.6959
## nodematch.Race.NatAm 0.9817 0.1842 0 5.329 <1e-04 ***
## nodematch.Race.Other -Inf 0.0000 0 -Inf <1e-04 ***
## nodematch.Race.White 1.2685 0.5371 0 2.362 0.0182 *
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Null Deviance: 28987 on 20910 degrees of freedom
## Residual Deviance: 1928 on 20898 degrees of freedom
##
## AIC: 1952 BIC: 2047 (Smaller is better.)
##
## Warning: The following terms have infinite coefficient estimates:
## nodematch.Race.Black nodematch.Race.Othermixingmatrix(mesa,"Race")## Note: Marginal totals can be misleading
## for undirected mixing matrices.
## Black Hisp NatAm Other White
## Black 0 8 13 0 5
## Hisp 8 53 41 1 22
## NatAm 13 41 46 0 10
## Other 0 1 0 0 0
## White 5 22 10 0 43.4.3.12 directed network
samplk3## Network attributes:
## vertices = 18
## directed = TRUE
## hyper = FALSE
## loops = FALSE
## multiple = FALSE
## bipartite = FALSE
## total edges= 56
## missing edges= 0
## non-missing edges= 56
##
## Vertex attribute names:
## cloisterville group vertex.names
##
## No edge attributesset.seed(1)
plot(samplk3)
3.4.3.13 Include mutual
sampmodel.01 <- ergm(samplk3~edges+mutual)## Starting maximum pseudolikelihood estimation (MPLE):## Evaluating the predictor and response matrix.## Maximizing the pseudolikelihood.## Finished MPLE.## Starting Monte Carlo maximum likelihood estimation (MCMLE):## Iteration 1 of at most 20:## Optimizing with step length 1.## The log-likelihood improved by 0.0002847.## Step length converged once. Increasing MCMC sample size.## Iteration 2 of at most 20:## Optimizing with step length 1.## The log-likelihood improved by 0.001122.## Step length converged twice. Stopping.## Finished MCMLE.## Evaluating log-likelihood at the estimate. Using 20 bridges: 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 .
## This model was fit using MCMC. To examine model diagnostics and check for degeneracy, use the mcmc.diagnostics() function.summary(sampmodel.01)##
## ==========================
## Summary of model fit
## ==========================
##
## Formula: samplk3 ~ edges + mutual
##
## Iterations: 2 out of 20
##
## Monte Carlo MLE Results:
## Estimate Std. Error MCMC % z value Pr(>|z|)
## edges -2.1505 0.2152 0 -9.993 <1e-04 ***
## mutual 2.2849 0.4684 0 4.878 <1e-04 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Null Deviance: 424.2 on 306 degrees of freedom
## Residual Deviance: 268.0 on 304 degrees of freedom
##
## AIC: 272 BIC: 279.5 (Smaller is better.)Strong mutual effect.
3.4.3.14 Include sender, receiver, mutual
p1 model
sampmodel.02 <- ergm(samplk3~ edges + sender + receiver + mutual)## Observed statistic(s) receiver10 are at their smallest attainable values. Their coefficients will be fixed at -Inf.## Starting maximum pseudolikelihood estimation (MPLE):## Evaluating the predictor and response matrix.## Maximizing the pseudolikelihood.## Finished MPLE.## Starting Monte Carlo maximum likelihood estimation (MCMLE):## Iteration 1 of at most 20:## Optimizing with step length 1.## The log-likelihood improved by 1.081.## Step length converged once. Increasing MCMC sample size.## Iteration 2 of at most 20:## Optimizing with step length 1.## The log-likelihood improved by 0.08992.## Step length converged twice. Stopping.## Finished MCMLE.## Evaluating log-likelihood at the estimate. Using 20 bridges: 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 .
## This model was fit using MCMC. To examine model diagnostics and check for degeneracy, use the mcmc.diagnostics() function.summary(sampmodel.02)##
## ==========================
## Summary of model fit
## ==========================
##
## Formula: samplk3 ~ edges + sender + receiver + mutual
##
## Iterations: 2 out of 20
##
## Monte Carlo MLE Results:
## Estimate Std. Error MCMC % z value Pr(>|z|)
## edges -2.315741 0.796090 0 -2.909 0.00363 **
## sender2 -0.438628 1.092996 0 -0.401 0.68819
## sender3 0.776450 1.093259 0 0.710 0.47757
## sender4 -0.008577 1.115291 0 -0.008 0.99386
## sender5 -0.452058 1.134888 0 -0.398 0.69039
## sender6 0.502989 1.095639 0 0.459 0.64617
## sender7 -0.252892 1.103350 0 -0.229 0.81871
## sender8 0.529555 1.111692 0 0.476 0.63382
## sender9 0.005037 1.122952 0 0.004 0.99642
## sender10 1.457053 1.055864 0 1.380 0.16760
## sender11 0.534553 1.100976 0 0.486 0.62730
## sender12 -0.446945 1.125660 0 -0.397 0.69133
## sender13 0.530122 1.107423 0 0.479 0.63215
## sender14 0.529945 1.117297 0 0.474 0.63528
## sender15 0.508301 1.123175 0 0.453 0.65087
## sender16 0.793735 1.081730 0 0.734 0.46309
## sender17 0.526922 1.107444 0 0.476 0.63422
## sender18 0.235750 1.103263 0 0.214 0.83079
## receiver2 0.772420 0.896048 0 0.862 0.38867
## receiver3 -0.739738 0.996962 0 -0.742 0.45809
## receiver4 0.017002 0.951789 0 0.018 0.98575
## receiver5 0.766819 0.911852 0 0.841 0.40038
## receiver6 -1.087262 1.114647 0 -0.975 0.32935
## receiver7 0.412868 0.901948 0 0.458 0.64713
## receiver8 -1.111177 1.098415 0 -1.012 0.31172
## receiver9 -0.001184 0.951954 0 -0.001 0.99901
## receiver10 -Inf 0.000000 0 -Inf < 1e-04 ***
## receiver11 -1.089252 1.082136 0 -1.007 0.31414
## receiver12 0.755936 0.907398 0 0.833 0.40480
## receiver13 -1.107780 1.098538 0 -1.008 0.31326
## receiver14 -1.092423 1.110361 0 -0.984 0.32519
## receiver15 -1.111820 1.099080 0 -1.012 0.31173
## receiver16 -2.004070 1.347086 0 -1.488 0.13683
## receiver17 -1.096807 1.109898 0 -0.988 0.32305
## receiver18 -0.480288 1.012901 0 -0.474 0.63538
## mutual 3.153694 0.636817 0 4.952 < 1e-04 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Null Deviance: 424.2 on 306 degrees of freedom
## Residual Deviance: NaN on 270 degrees of freedom
##
## AIC: NaN BIC: NaN (Smaller is better.)
##
## Warning: The following terms have infinite coefficient estimates:
## receiver10sna::degree(samplk3,cmode = "indegree")## [1] 4 6 3 4 6 2 5 2 4 0 2 6 2 2 2 1 2 33.4.3.15 Other ergm-terms
Details of other ergm-terms see https://www.jstatsoft.org/article/view/v024i04.
3.4.4 Missing edges
Make sure to set missing edges as NA: probability of an edge in the observed sample.
missnet=flomarriage
missnet[1,2]=missnet[2,4]=missnet[5,6]=NA # originally are 0
summary(ergm(missnet~edges))## Starting maximum pseudolikelihood estimation (MPLE):## Evaluating the predictor and response matrix.## Maximizing the pseudolikelihood.## Finished MPLE.## Stopping at the initial estimate.## Evaluating log-likelihood at the estimate.##
## ==========================
## Summary of model fit
## ==========================
##
## Formula: missnet ~ edges
##
## Iterations: 5 out of 20
##
## Monte Carlo MLE Results:
## Estimate Std. Error MCMC % z value Pr(>|z|)
## edges -1.5790 0.2456 0 -6.43 <1e-04 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Null Deviance: 162.2 on 117 degrees of freedom
## Residual Deviance: 107.0 on 116 degrees of freedom
##
## AIC: 109 BIC: 111.8 (Smaller is better.)summary(ergm(flomarriage~edges))## Starting maximum pseudolikelihood estimation (MPLE):
## Evaluating the predictor and response matrix.
## Maximizing the pseudolikelihood.
## Finished MPLE.
## Stopping at the initial estimate.
## Evaluating log-likelihood at the estimate.##
## ==========================
## Summary of model fit
## ==========================
##
## Formula: flomarriage ~ edges
##
## Iterations: 5 out of 20
##
## Monte Carlo MLE Results:
## Estimate Std. Error MCMC % z value Pr(>|z|)
## edges -1.6094 0.2449 0 -6.571 <1e-04 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Null Deviance: 166.4 on 120 degrees of freedom
## Residual Deviance: 108.1 on 119 degrees of freedom
##
## AIC: 110.1 BIC: 112.9 (Smaller is better.)#Set NA
1/(1+exp(-(-1.5790)))## [1] 0.170937220/(choose(16,2)-3)## [1] 0.1709402#Set 0
1/(1+exp(-(-1.6094)))## [1] 0.166671920/(choose(16,2))## [1] 0.16666673.5 simulate netowrks from an ergm model fit
The ergm model defines a probability distibution across all networks of this size(#nodes fixed).
Use simulate(ergm-model,nsim=n) to get a list of networks
Use summary and functions that can be used to a list
summary(flomodel.04)##
## ==========================
## Summary of model fit
## ==========================
##
## Formula: flomarriage ~ edges + absdiff("wealth")
##
## Iterations: 4 out of 20
##
## Monte Carlo MLE Results:
## Estimate Std. Error MCMC % z value Pr(>|z|)
## edges -2.302042 0.401906 0 -5.728 <1e-04 ***
## absdiff.wealth 0.015519 0.006157 0 2.521 0.0117 *
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Null Deviance: 166.4 on 120 degrees of freedom
## Residual Deviance: 102.0 on 118 degrees of freedom
##
## AIC: 106 BIC: 111.5 (Smaller is better.)flomodel.04.sim <- simulate(flomodel.04, nsim=10,seed = 1)## Warning: You appear to be calling simulate.formula() directly.
## simulate.formula() is a method, and will not be exported in a future
## version of 'ergm'. Use simulate() instead, or getS3method() if absolutely
## necessary.class(flomodel.04.sim)## [1] "network.list"summary(flomodel.04.sim)## Number of Networks: 10
## Model: flomarriage ~ edges + absdiff("wealth")
## Reference: ~Bernoulli
## Constraints: ~.
## Parameters:
## edges absdiff.wealth
## -2.3020421 0.0155192
##
## Stored network statistics:
## edges absdiff.wealth
## [1,] 20 1070
## [2,] 22 1170
## [3,] 19 734
## [4,] 18 1123
## [5,] 18 1215
## [6,] 15 697
## [7,] 20 1165
## [8,] 31 1577
## [9,] 19 1138
## [10,] 22 1136## Number of Networks: 10
## Model: flomarriage ~ edges + absdiff("wealth")
## Reference: ~Bernoulli
## Constraints: ~.
## Parameters:
## edges absdiff.wealth
## -2.3020421 0.0155192length(flomodel.04.sim)## [1] 10flomodel.04.sim[[4]]## Network attributes:
## vertices = 16
## directed = FALSE
## hyper = FALSE
## loops = FALSE
## multiple = FALSE
## bipartite = FALSE
## total edges= 18
## missing edges= 0
## non-missing edges= 18
##
## Vertex attribute names:
## priorates totalties vertex.names wealth
##
## No edge attributesplot(flomodel.04.sim[[4]], label= flomodel.04.sim[[4]] %v% "vertex.names")
flomarriage## Network attributes:
## vertices = 16
## directed = FALSE
## hyper = FALSE
## loops = FALSE
## multiple = FALSE
## bipartite = FALSE
## total edges= 20
## missing edges= 0
## non-missing edges= 20
##
## Vertex attribute names:
## priorates totalties vertex.names wealth
##
## No edge attributesformula: Specify your own model
basis: initial network for MCcontrol: settings of MCMC
basenet <- network(16,density=0.1,directed=FALSE)
g.sim <- simulate(flomarriage ~ edges + kstar(2), nsim=1000, coef=c(-1.8,0.03),
basis=basenet, control=control.simulate(MCMC.burnin=1000, MCMC.interval=10))
# MCMC.interval: Number of proposals between sampled statistics.
length(g.sim)## [1] 1000?simulate.ergm3.6 Check the goodness of fit
- AIC,BIC; Deviance;
- Goodness of fit:
gof - MCMC diagnostics:
mcmc.diagnostics
3.6.1 gof
gof(model~model+degree+esp+distance)
Only four possible arguments for ergm-term:
model: all the terms included in the modeldegree: degree (node level)esp: edgwise share partners (edge level)distance: geodesic distances (dyad level)
Draw samples from the specified model, calculate the MC p-value based on the distribution generated by the samples.
flo.04.gof.model=gof(flomodel.04~model+degree+esp+distance)
flo.04.gof.model##
## Goodness-of-fit for model statistics
##
## obs min mean max MC p-value
## edges 20 12 20.22 30 1.00
## absdiff.wealth 1146 565 1142.44 1841 0.96
##
## Goodness-of-fit for degree
##
## obs min mean max MC p-value
## 0 1 0 1.24 5 1.00
## 1 4 0 3.15 8 0.74
## 2 2 0 4.51 9 0.24
## 3 6 0 3.32 7 0.18
## 4 2 0 2.01 6 1.00
## 5 0 0 0.91 4 0.72
## 6 1 0 0.50 3 0.82
## 7 0 0 0.22 2 1.00
## 8 0 0 0.10 1 1.00
## 9 0 0 0.02 1 1.00
## 10 0 0 0.02 1 1.00
##
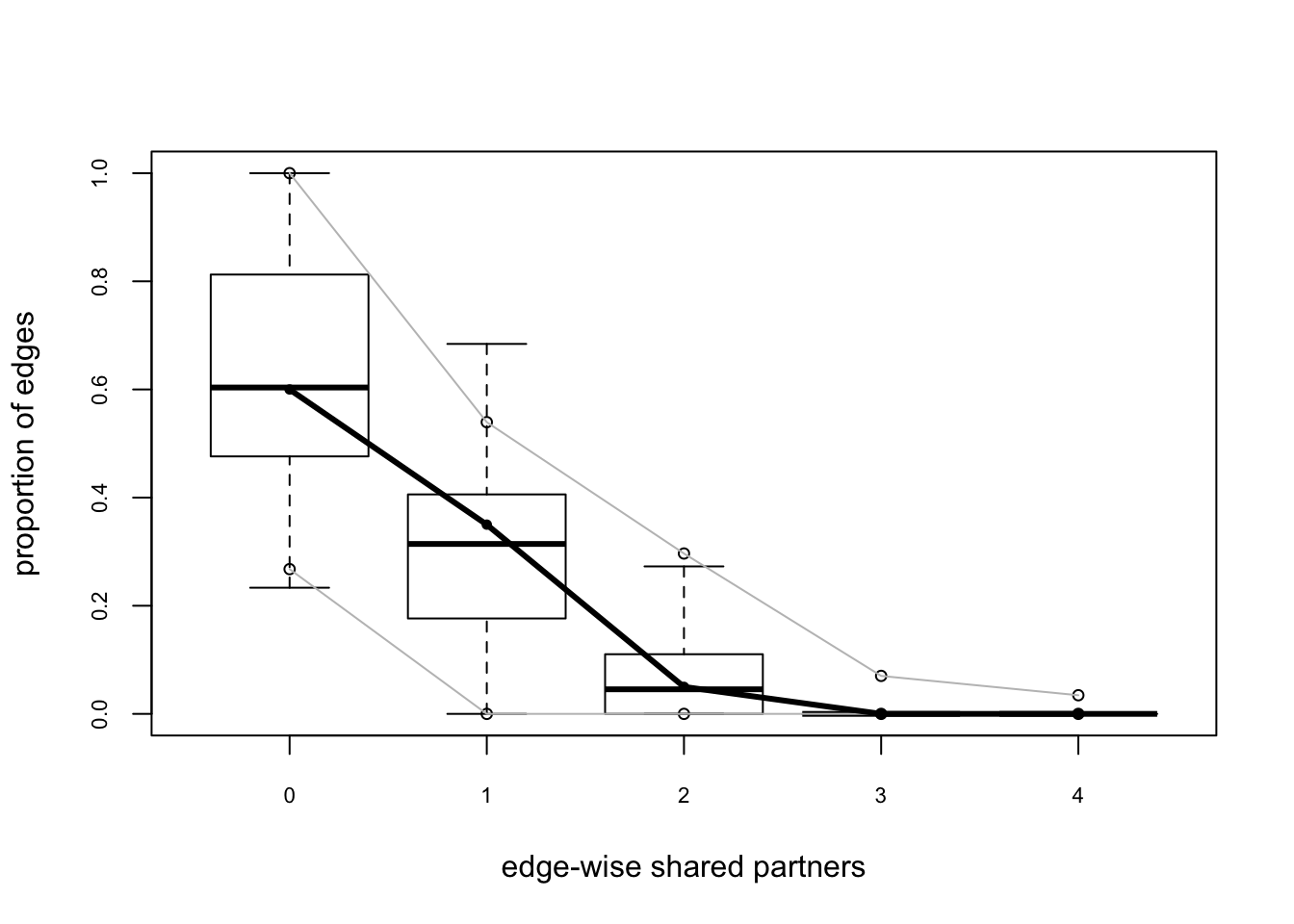
## Goodness-of-fit for edgewise shared partner
##
## obs min mean max MC p-value
## esp0 12 5 12.39 23 1.00
## esp1 7 0 5.99 14 0.94
## esp2 1 0 1.60 12 1.00
## esp3 0 0 0.19 4 1.00
## esp4 0 0 0.05 1 1.00
##
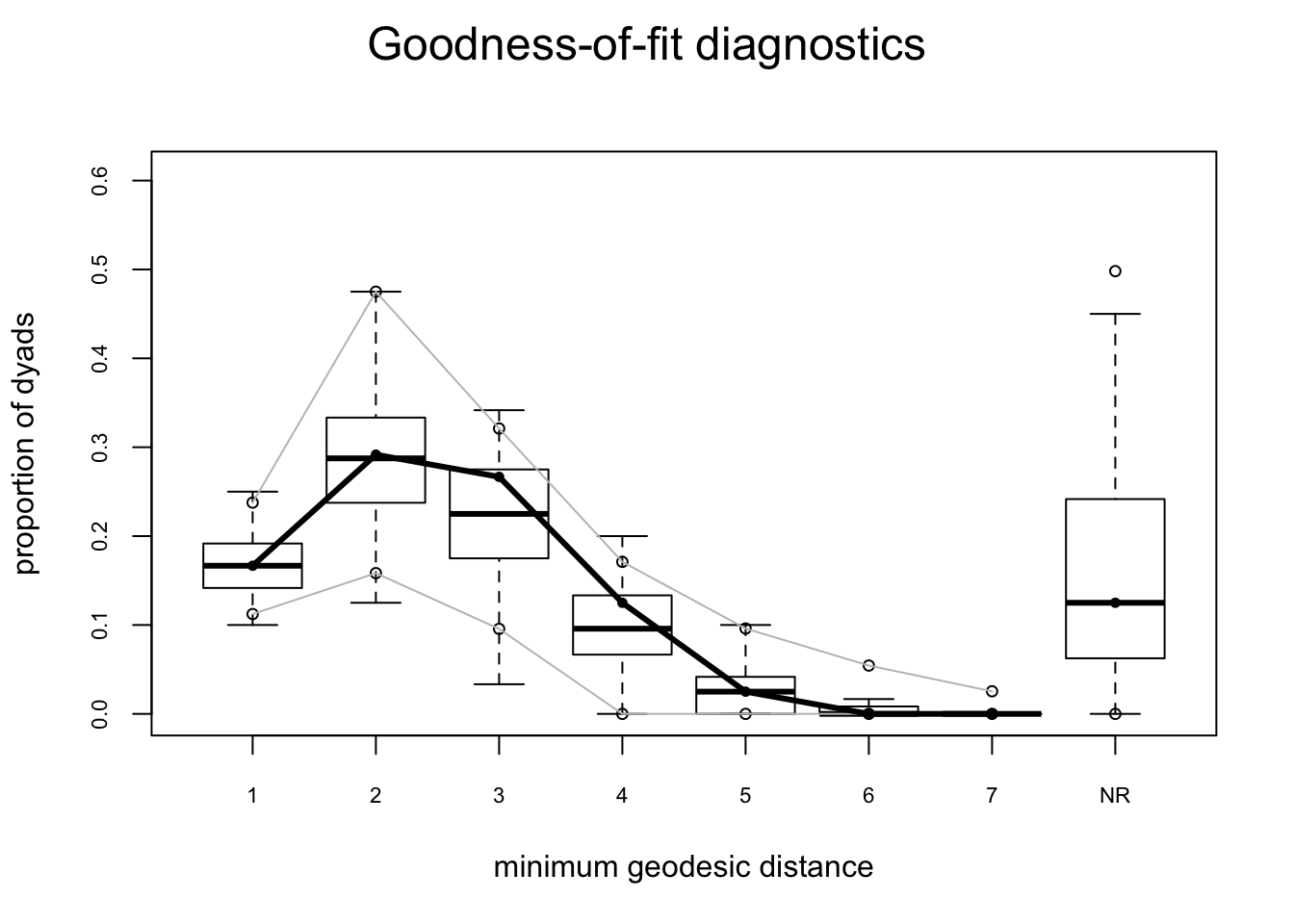
## Goodness-of-fit for minimum geodesic distance
##
## obs min mean max MC p-value
## 1 20 12 20.22 30 1.00
## 2 35 15 35.01 58 1.00
## 3 32 4 26.70 41 0.58
## 4 15 0 11.11 24 0.60
## 5 3 0 3.35 14 1.00
## 6 0 0 0.89 11 1.00
## 7 0 0 0.21 6 1.00
## 8 0 0 0.06 3 1.00
## 9 0 0 0.02 2 1.00
## Inf 15 0 22.43 73 1.00names(flo.04.gof.model)## [1] "network.size" "GOF" "pval.model" "summary.model"
## [5] "pobs.model" "psim.model" "bds.model" "obs.model"
## [9] "sim.model" "pval.dist" "summary.dist" "pobs.dist"
## [13] "psim.dist" "bds.dist" "obs.dist" "sim.dist"
## [17] "pval.deg" "summary.deg" "pobs.deg" "psim.deg"
## [21] "bds.deg" "obs.deg" "sim.deg" "pval.espart"
## [25] "summary.espart" "pobs.espart" "psim.espart" "bds.espart"
## [29] "obs.espart" "sim.espart"#95% confidence interval
plot(flo.04.gof.model)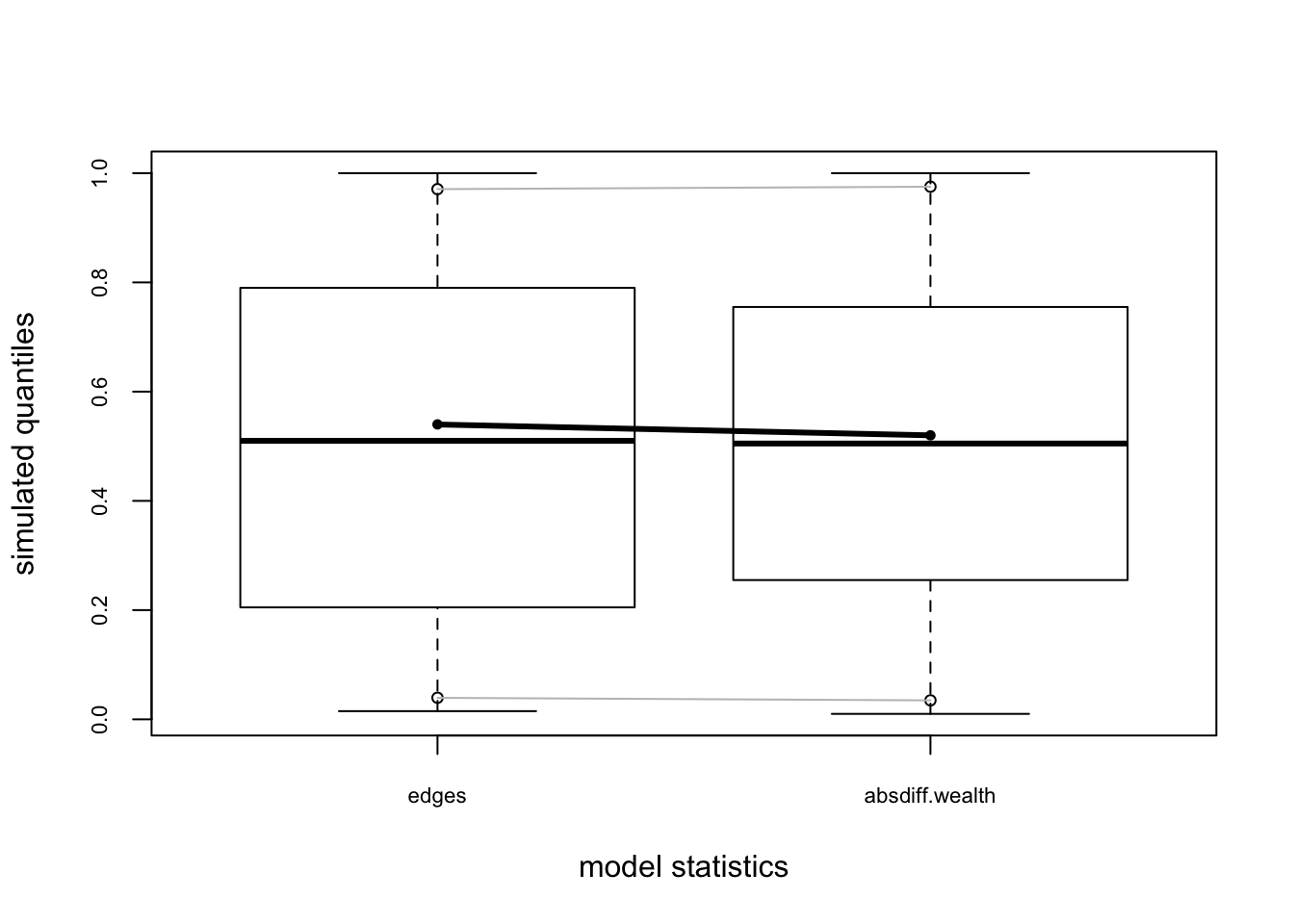
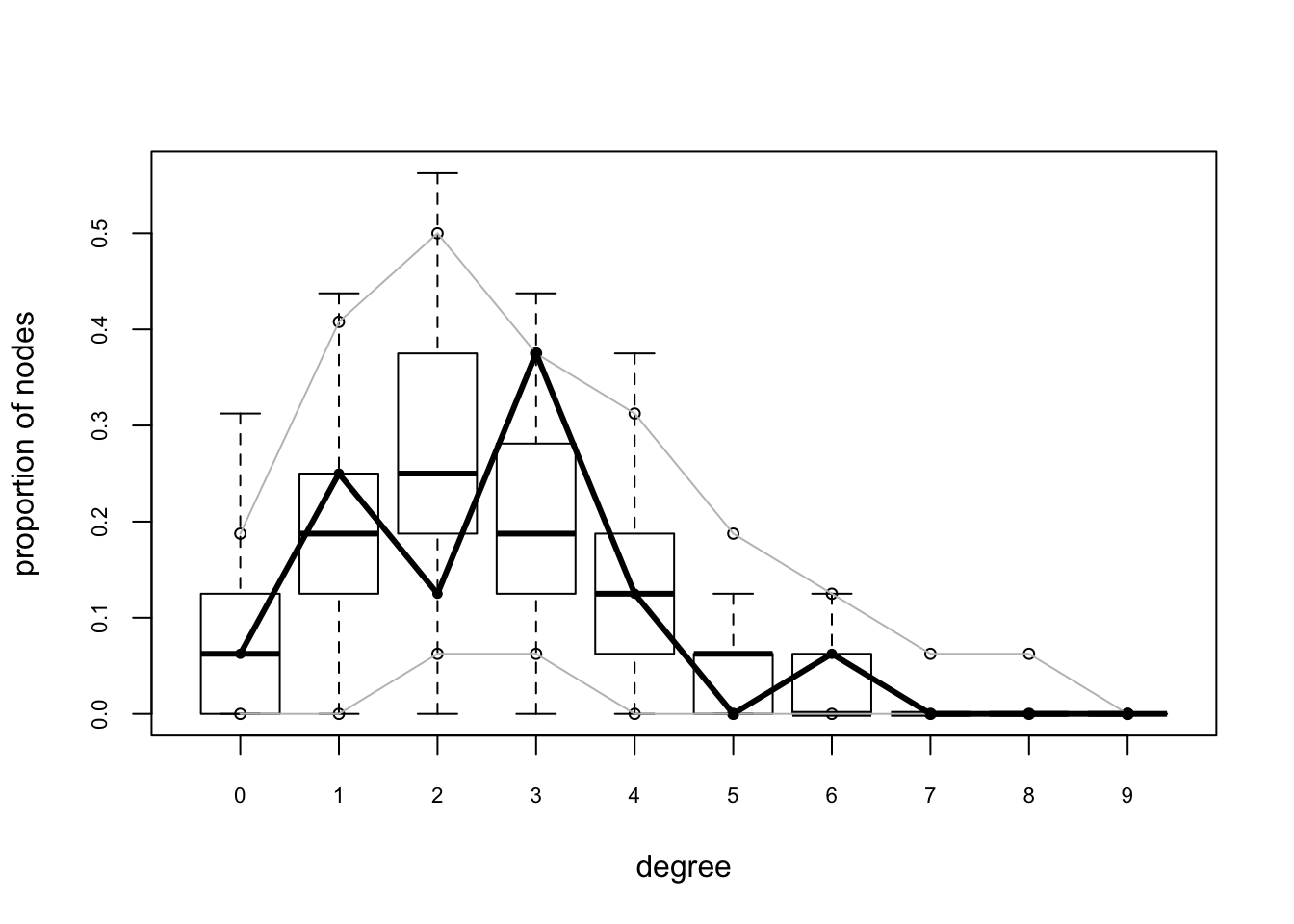
3.6.2 mcmc.diagnostics
Check the MCMC
#install.packages("latticeExtra")
fit <- ergm(flobusiness ~ edges+degree(1))## Starting maximum pseudolikelihood estimation (MPLE):## Evaluating the predictor and response matrix.## Maximizing the pseudolikelihood.## Finished MPLE.## Starting Monte Carlo maximum likelihood estimation (MCMLE):## Iteration 1 of at most 20:## Optimizing with step length 1.## The log-likelihood improved by 0.2425.## Step length converged once. Increasing MCMC sample size.## Iteration 2 of at most 20:## Optimizing with step length 1.## The log-likelihood improved by 0.001113.## Step length converged twice. Stopping.## Finished MCMLE.## Evaluating log-likelihood at the estimate. Using 20 bridges: 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 .
## This model was fit using MCMC. To examine model diagnostics and check for degeneracy, use the mcmc.diagnostics() function.mcmc.diagnostics(fit) #centered## Sample statistics summary:
##
## Iterations = 16384:4209664
## Thinning interval = 1024
## Number of chains = 1
## Sample size per chain = 4096
##
## 1. Empirical mean and standard deviation for each variable,
## plus standard error of the mean:
##
## Mean SD Naive SE Time-series SE
## edges -0.12183 3.766 0.05884 0.05884
## degree1 0.07349 1.634 0.02553 0.02553
##
## 2. Quantiles for each variable:
##
## 2.5% 25% 50% 75% 97.5%
## edges -7 -3 0 2 7
## degree1 -4 -1 0 1 3
##
##
## Sample statistics cross-correlations:
## edges degree1
## edges 1.0000 -0.4341
## degree1 -0.4341 1.0000
##
## Sample statistics auto-correlation:
## Chain 1
## edges degree1
## Lag 0 1.000000000 1.000000000
## Lag 1024 0.006760343 0.001466041
## Lag 2048 0.008602875 -0.006057681
## Lag 3072 -0.018218817 -0.011568388
## Lag 4096 0.028773889 -0.015764465
## Lag 5120 -0.015302900 -0.004412151
##
## Sample statistics burn-in diagnostic (Geweke):
## Chain 1
##
## Fraction in 1st window = 0.1
## Fraction in 2nd window = 0.5
##
## edges degree1
## 0.9156 -0.5825
##
## Individual P-values (lower = worse):
## edges degree1
## 0.3598522 0.5602179
## Joint P-value (lower = worse): 0.6503828 .## Warning in formals(fun): argument is not a function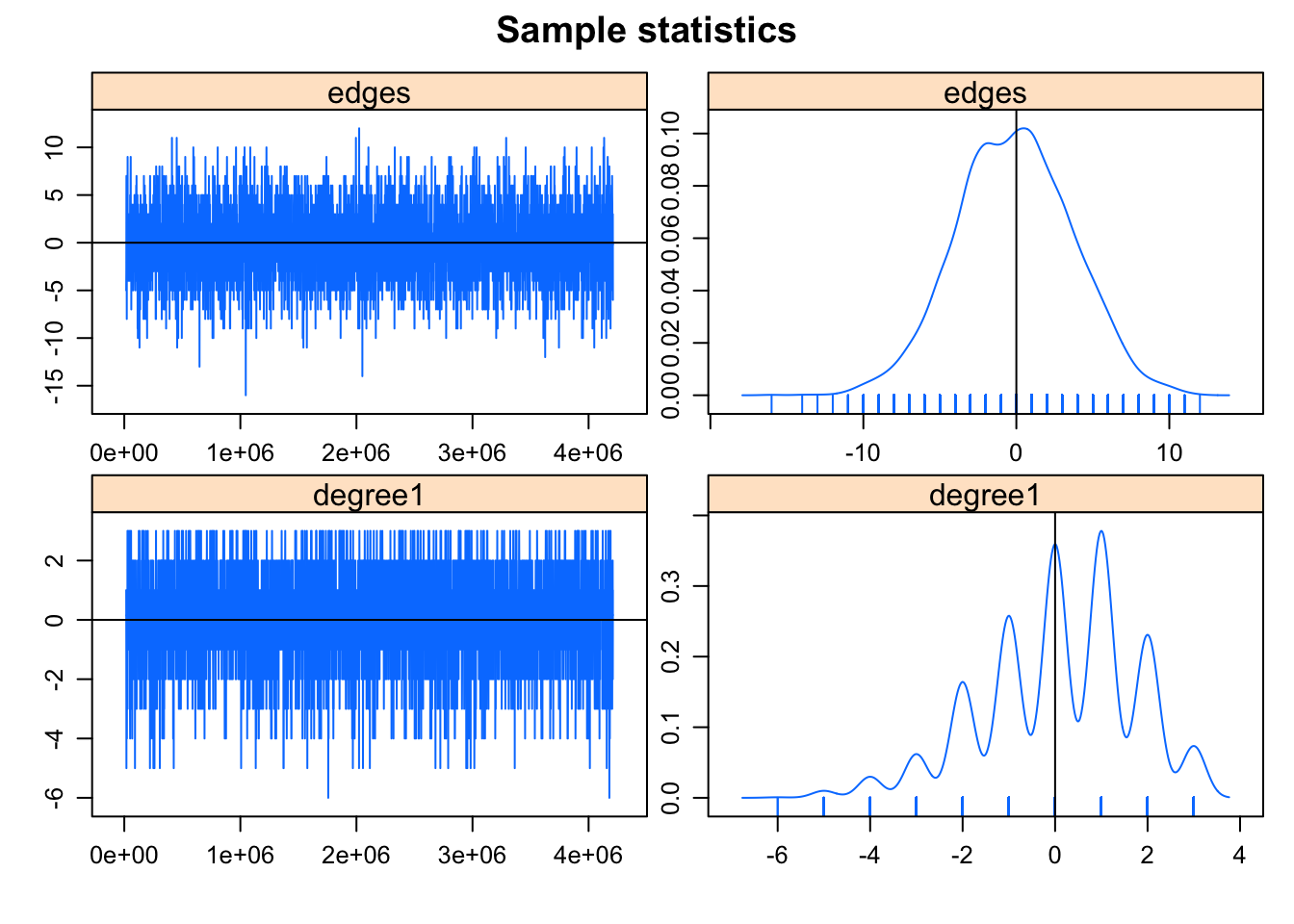
##
## MCMC diagnostics shown here are from the last round of simulation, prior to computation of final parameter estimates. Because the final estimates are refinements of those used for this simulation run, these diagnostics may understate model performance. To directly assess the performance of the final model on in-model statistics, please use the GOF command: gof(ergmFitObject, GOF=~model).Interpretation on mcmc.diagnostics:
Good example.
- Mixing well: MCMC sample statistics are varying randomly around the observed values at each step
- Bell-shaped centered at 0. The difference between the observed and simulated values of the sample statistics have a roughly bell-shaped distribution, centered at 0.
- Notice the sawtooth pattern is due to the discrete values. The sawtooth pattern visible on the degree term deviation plot is due to the combination of discrete values and small range in the statistics: the observed number of degree 1 nodes is 3, and only a few discrete values are produced by the simulations.
Bad example:
](images/mcdiag.png)
Figure 3.1: Example from Tutorial
3.7 More functions
](images/more.png)
Figure 3.2: Example from Tutorial