Data Wrangling
STA 210 - Summer 2022
Yunran Chen
Welcome
Topics
Data Cleaning/Wrangling
Course Evaluation (10-15min)
Data Cleaning using tidyverse
Raw data to understanding, insight, and knowledge
Workflow for real-world data analysis
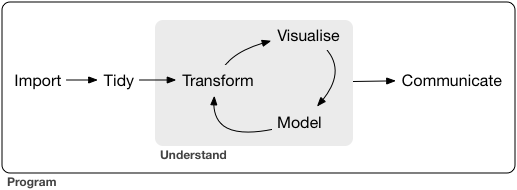
Packages in Tidyverse
Data Wrangling
Focus on data wrangling
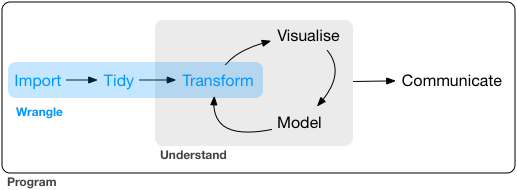
Data import (
readr,tibble)Tidy data (
tidyr)Wrangle (
dplyr,stringr,lubridate,janitor)
Data import
Data import using readr
read_csv, …
Extract the certain type of data
readr::parse_*: parse the characters/numbers only

function parse in pkg readr
[1] 100[1] 20[1] 123.45[1] 123456789[1] 123456789[1] 123456789function parse in pkg readr
function parse in pkg readr
[1] "2015-01-01"[1] "2015-01-02"[1] "2015-02-01"[1] "2001-02-15"Other packages for data importing
Package
haven: SPSS, Stata, SAS filePackage
readxl: Excel file .xls, .xlsxPackage
jsonlite/htmltab: json, htmluse
as_tibbleto coerce a data frame to a tibble
janitor package can help with cleaning names
clean_names,remove_empty_cols,remove_empty_rows

janitor package
clean_names,remove_empty_cols,remove_empty_rows
Tidy data
Tidy data

Tidy data
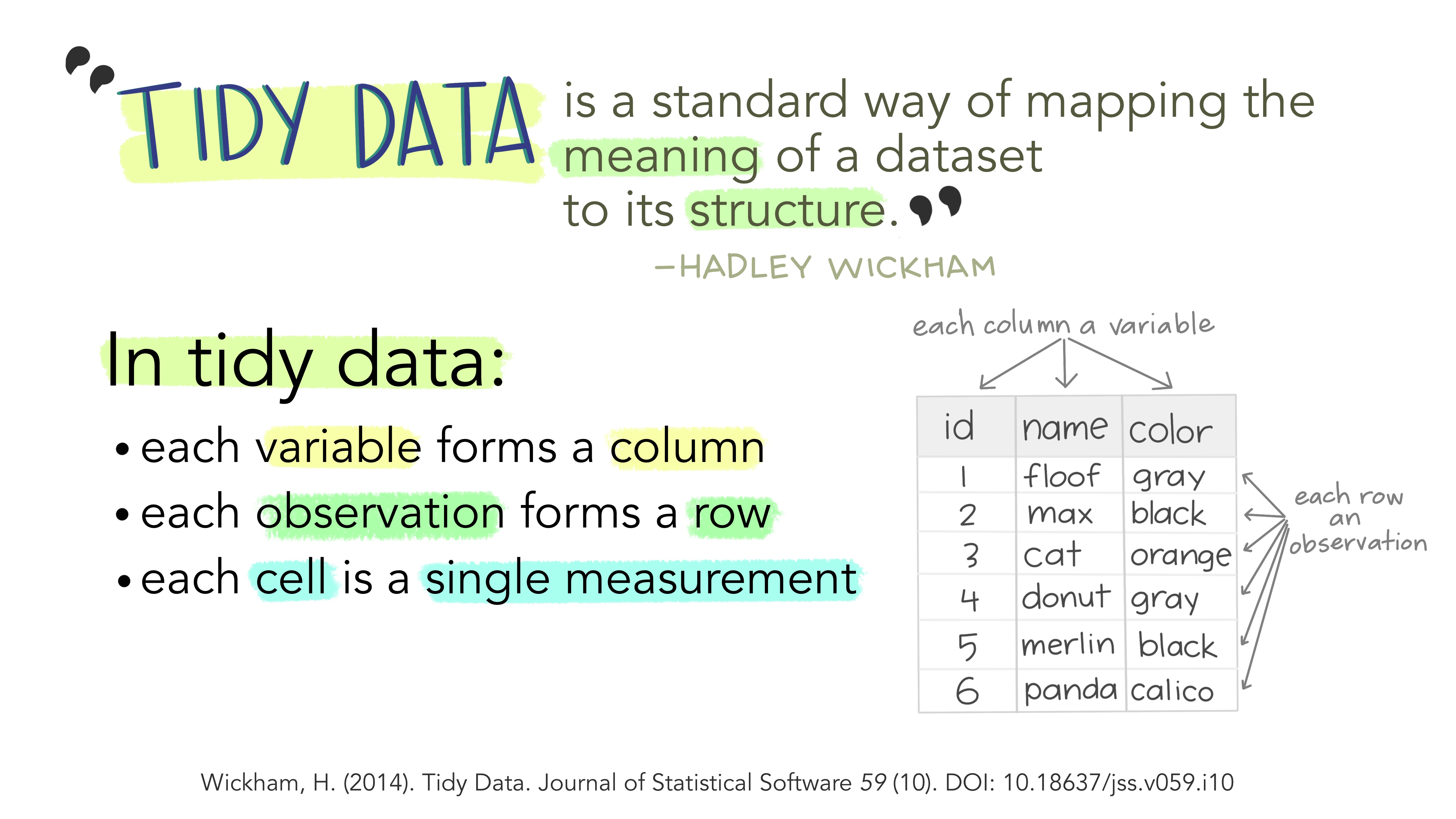
Tidy data
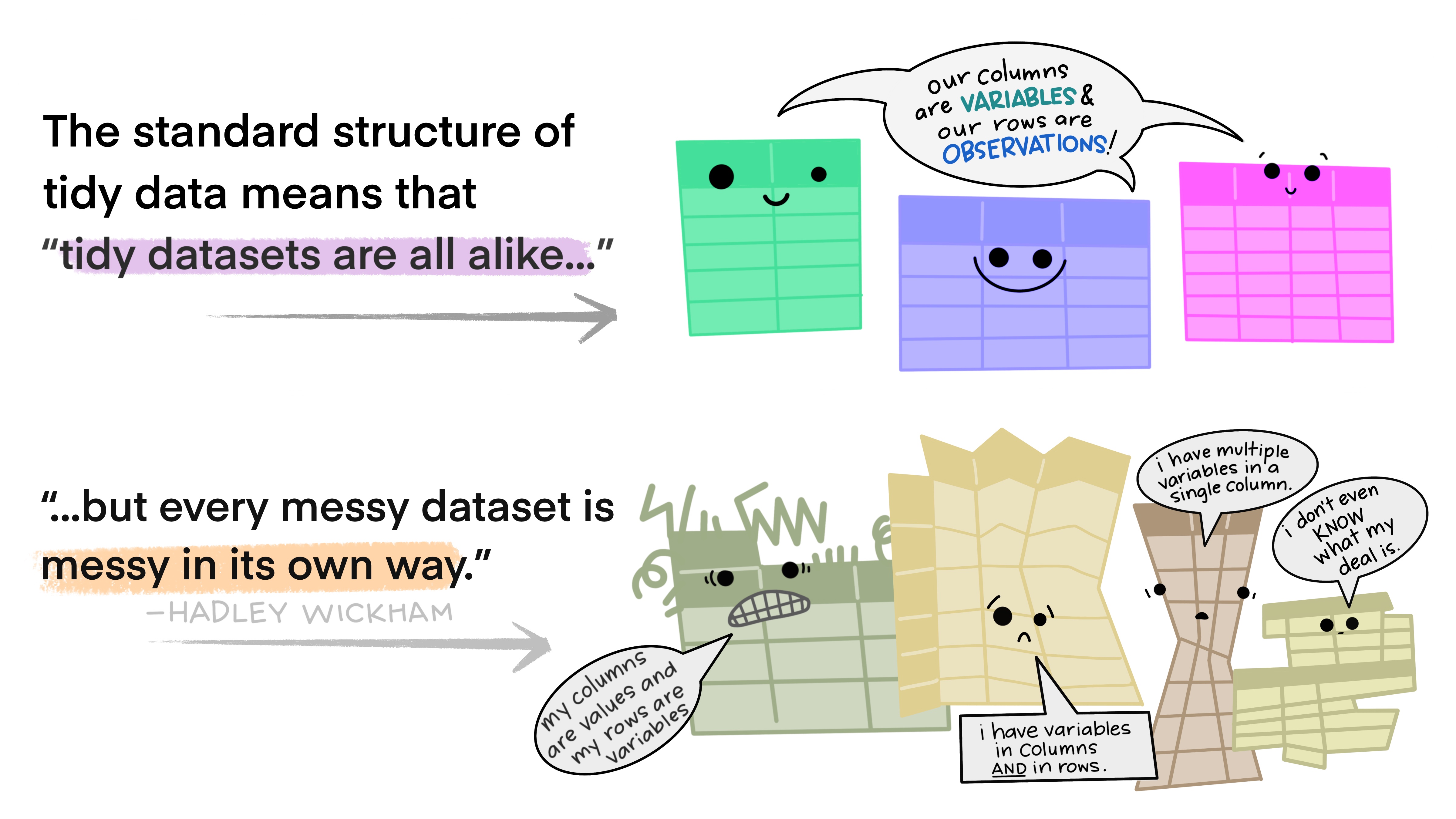
Tidy data
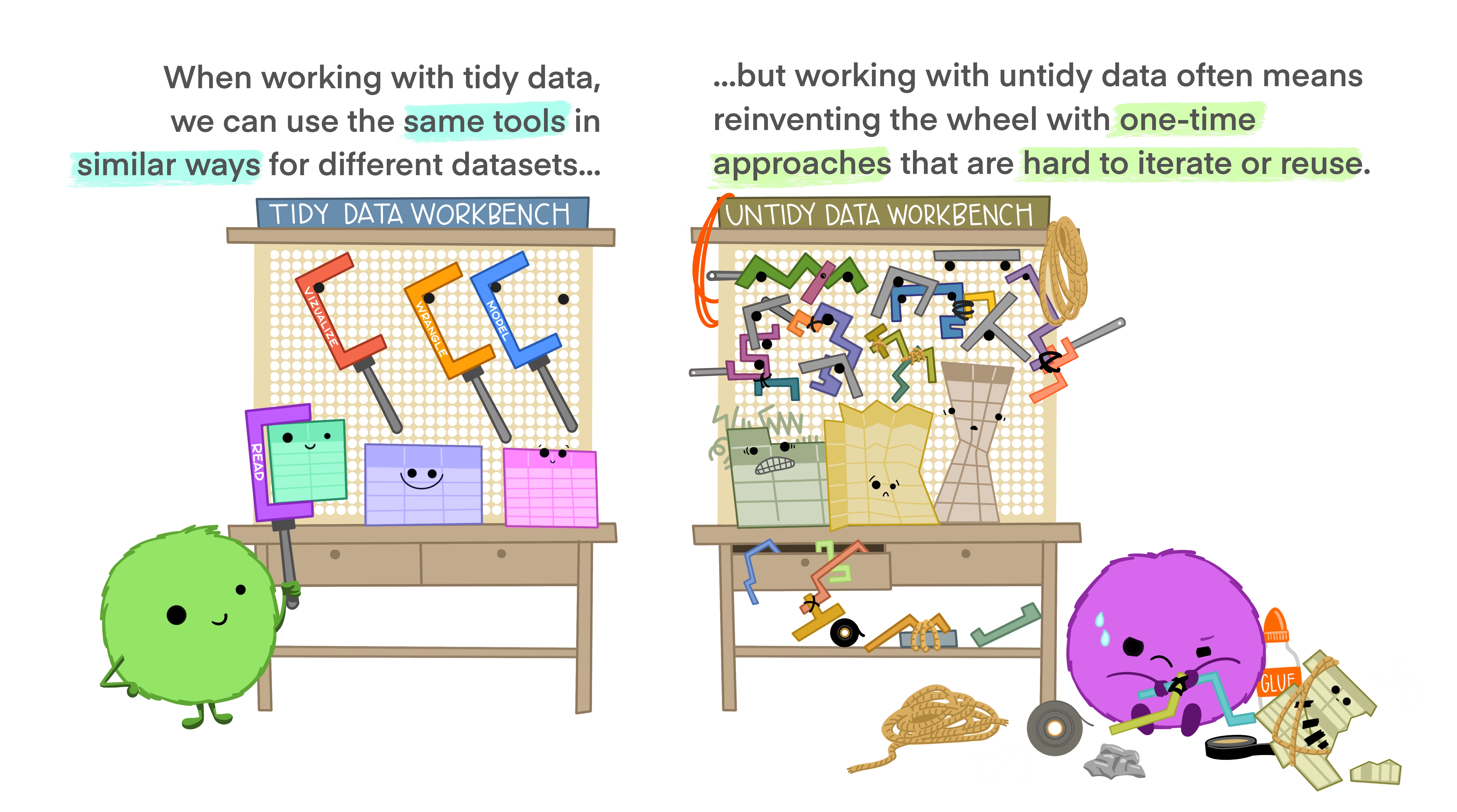
Tidy data
Tidy data

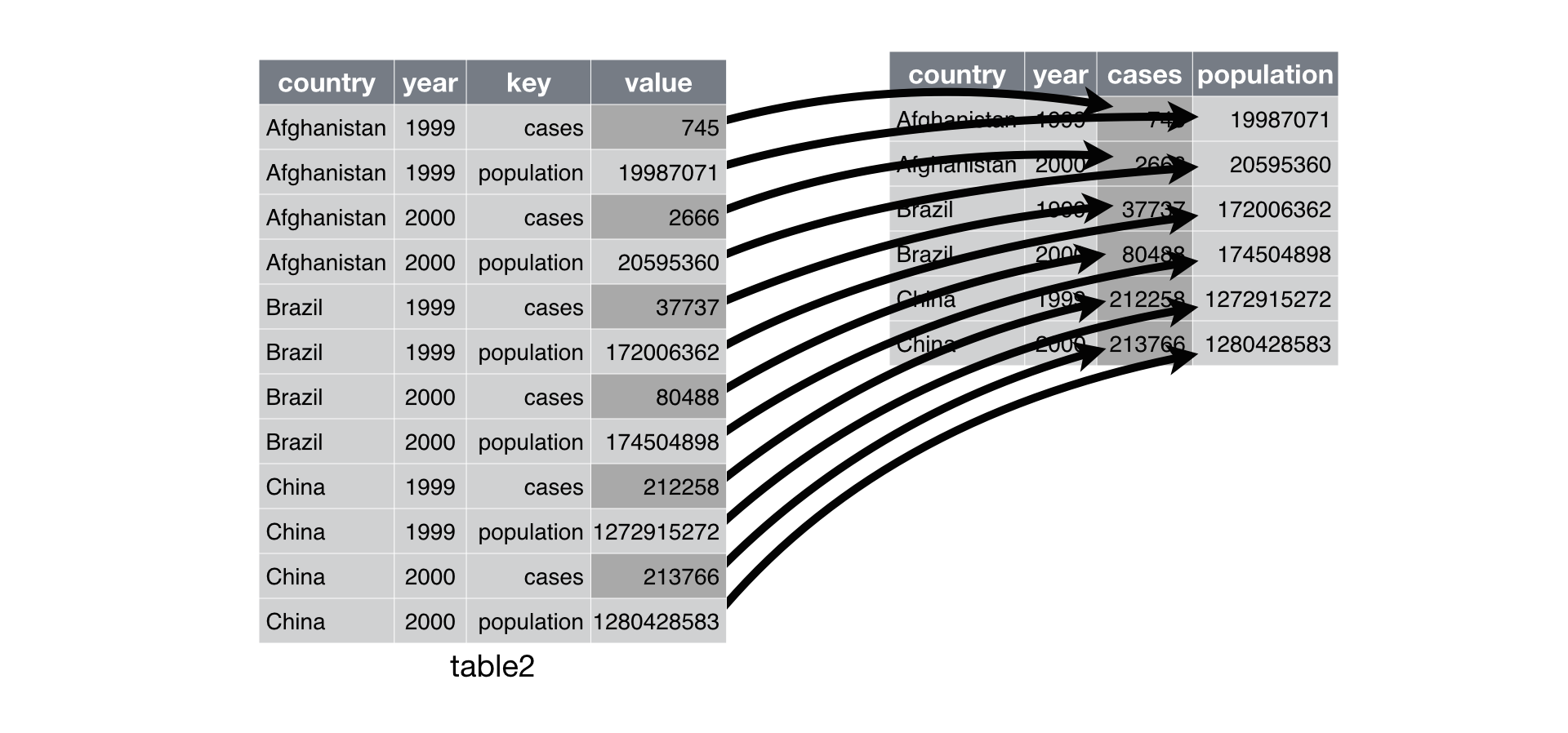
pivot_longer function in tidyr pkg
pivot_longer: from wide to long
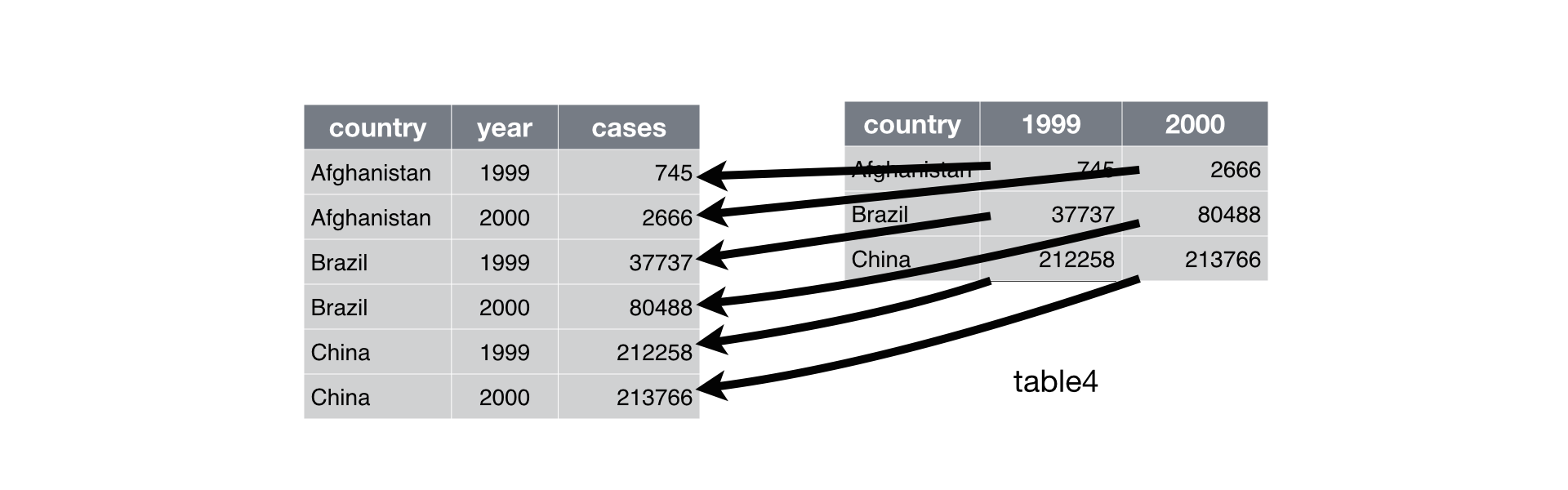
pivot_wider function in tidyr pkg
pivot_wider: from long to wide
separate function in tidyr pkg
separate: from 1 column to 2+ column- can
sepbased on digits or characters.
unite function in tidyr pkg
unite: from 2+ column to 1 column
Transform
Packages in Tidyverse

dplyr: join multiple datasets
Relational datasets in nycflights13
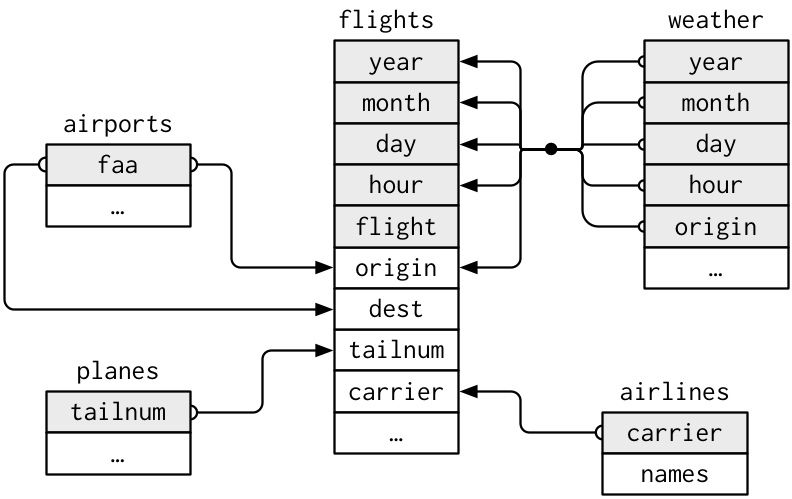
join multiple datasets
R pkg nycflights13 provide multiple relational datasets:
flightsconnects toplanesvia a single variable,tailnum.flightsconnects toairlinesthrough thecarriervariable.flightsconnects toairportsin two ways: via theoriginanddestvariables.flightsconnects toweatherviaorigin(the location), andyear,month,dayandhour(the time)
inner_join
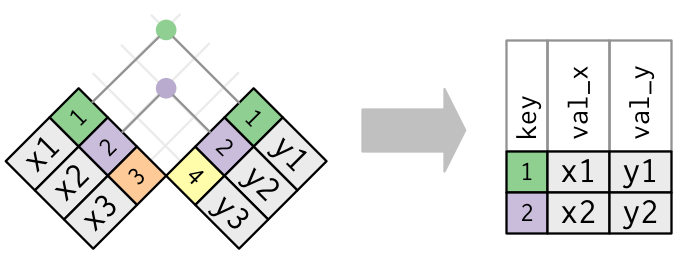
*_join
left_join, right_join, full_join
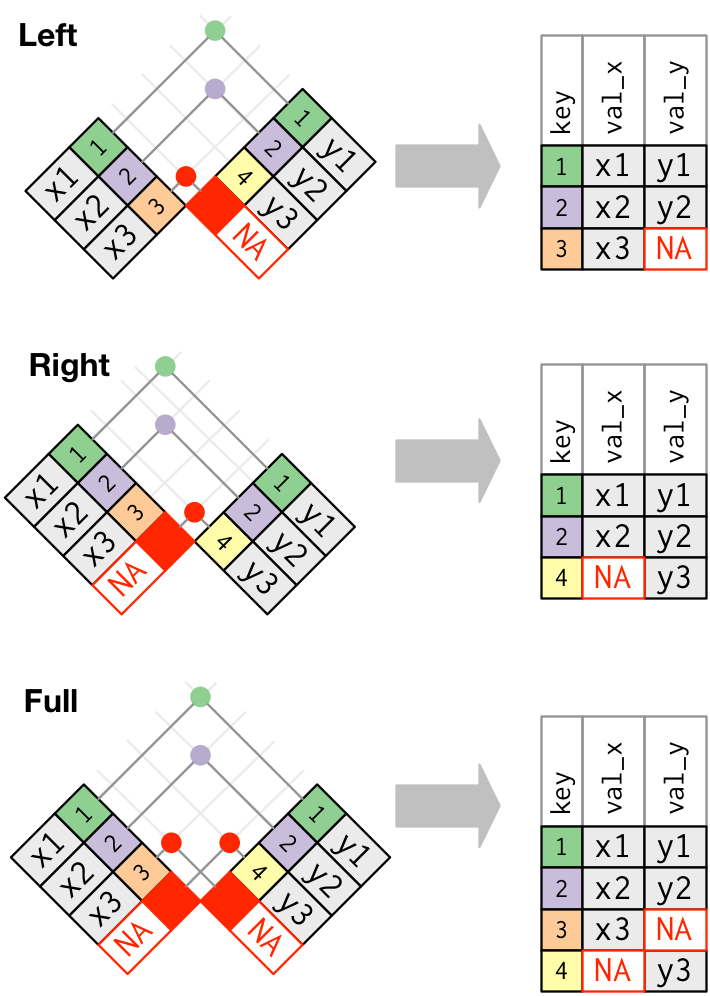
Data wrangling with dplyr
slice(): pick rows using indexesfilter(): keep rows satisfying your conditionselect(): select variables obtain a tibblepull(): grab a column as a vectorrelocate(): relocate a variablearrange(): reorder rowsrename(): rename a variablemutate(): add columnsgroup_by()%>%summarize(): summary statistics for different groupscount(): count the frequencydistinct(): keep unique rows- functions within
mutate():across(),if_else(),case_when() - functions for selecting variables:
starts_with(),ends_with(),contains(),matches(),everything()
case_when

Practice
- Keep the chinstrap and gentoo penguins, living in Dream and Biscoe Islands.
- Get first 100 observation
- Only keep columns from
speciestoflipper_length_mm, andsexandyear - Rename
sexasgender - Move
genderright after island, move numeric variables after factor variables - Add a new column to identify each observation
- Transfer
islandas character - Add a new variable called
bill_ratiowhich is the ratio of bill length to bill depth - Obtain the mean and standard deviation of body mass of different species
- For different species, obtain the mean of variables ending with
mm - Provide the distribution of different species of penguins living in different island across time
Practice
To penguins, add a new column size_bin that contains:
- “large” if body mass is greater than 4500 g
- “medium” if body mass is greater than 3000 g, and less than or equal to 4500 g
- “small” if body mass is less than or equal to 3000 g
Deal with different types of variables
Deal with different types of variables
stringrfor stringsforcatsfor factorslubridatefor dates and times
stringr for strings
stringr for strings
Useful functions in stringr
If your raw data has numbers as variable names, you may consider to add characters in front of it.
Useful functions in stringr
To obtain nice variable names. - Can use janitor::clean_names - Or str_to_upper, str_to_lower, str_to_title
Useful functions in stringr
str_detect(): Return TRUE/FALSE for strings satisfying the pattern- pattern: regular expressions (CheatSheets)
str_replace,str_replace_all(multiple replacements)- Text mining: Useful if your have text in the survey, and you want to extract important features from text
forcats for factors
forcats for factors
fct_infreq: order levels by frequencyfct_rev: reverse the order of levelsfct_reorder,fct_reorder2: order according to other variablesfct_relevel: reorder manuallyfct_collapse: combine levels- Useful for visualization in
ggplot.
fct_infreq and fct_rev
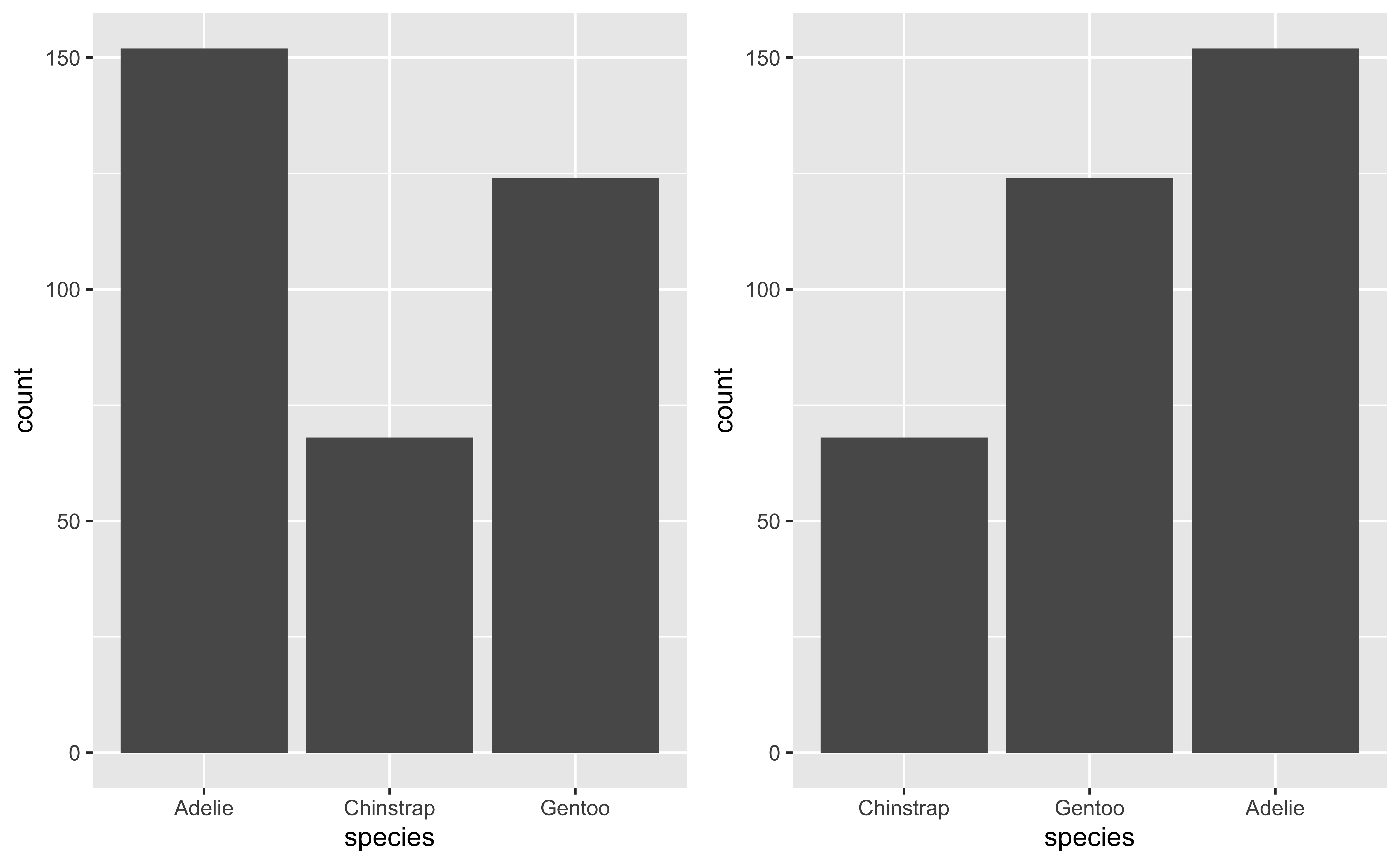
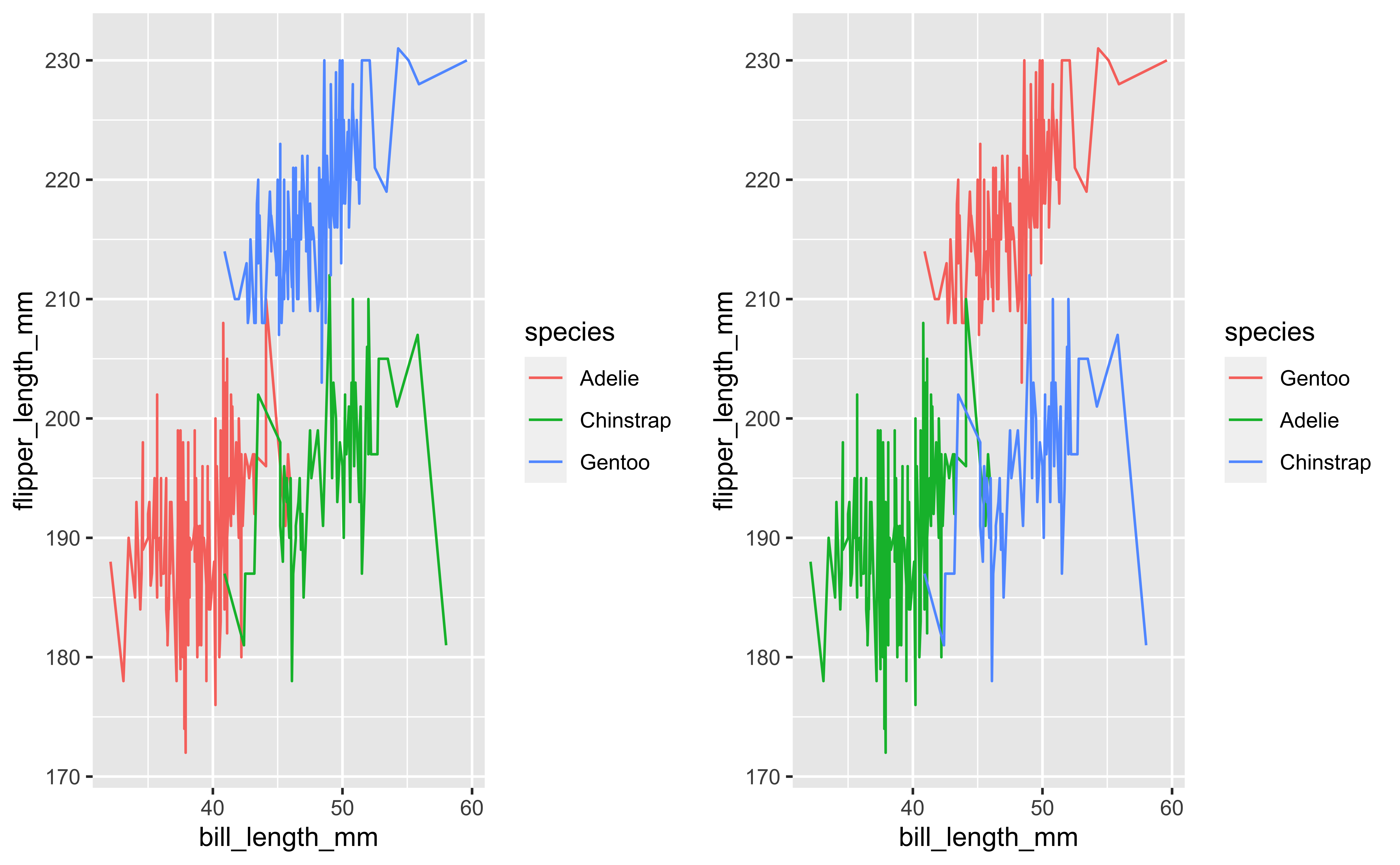
Examples for fct_reorder
library(cowplot)
peng_summary=penguins %>%
group_by(species)%>%
summarise(
bill_length_mean=mean(bill_length_mm, na.rm=T),
bill_depth_mean=mean(bill_depth_mm, na.rm=T))
p1=ggplot(peng_summary,aes(bill_length_mean,species)) +
geom_point()
p2=ggplot(peng_summary,aes(bill_length_mean,fct_reorder(species,bill_length_mean))) + geom_point()
plot_grid(p1,p2)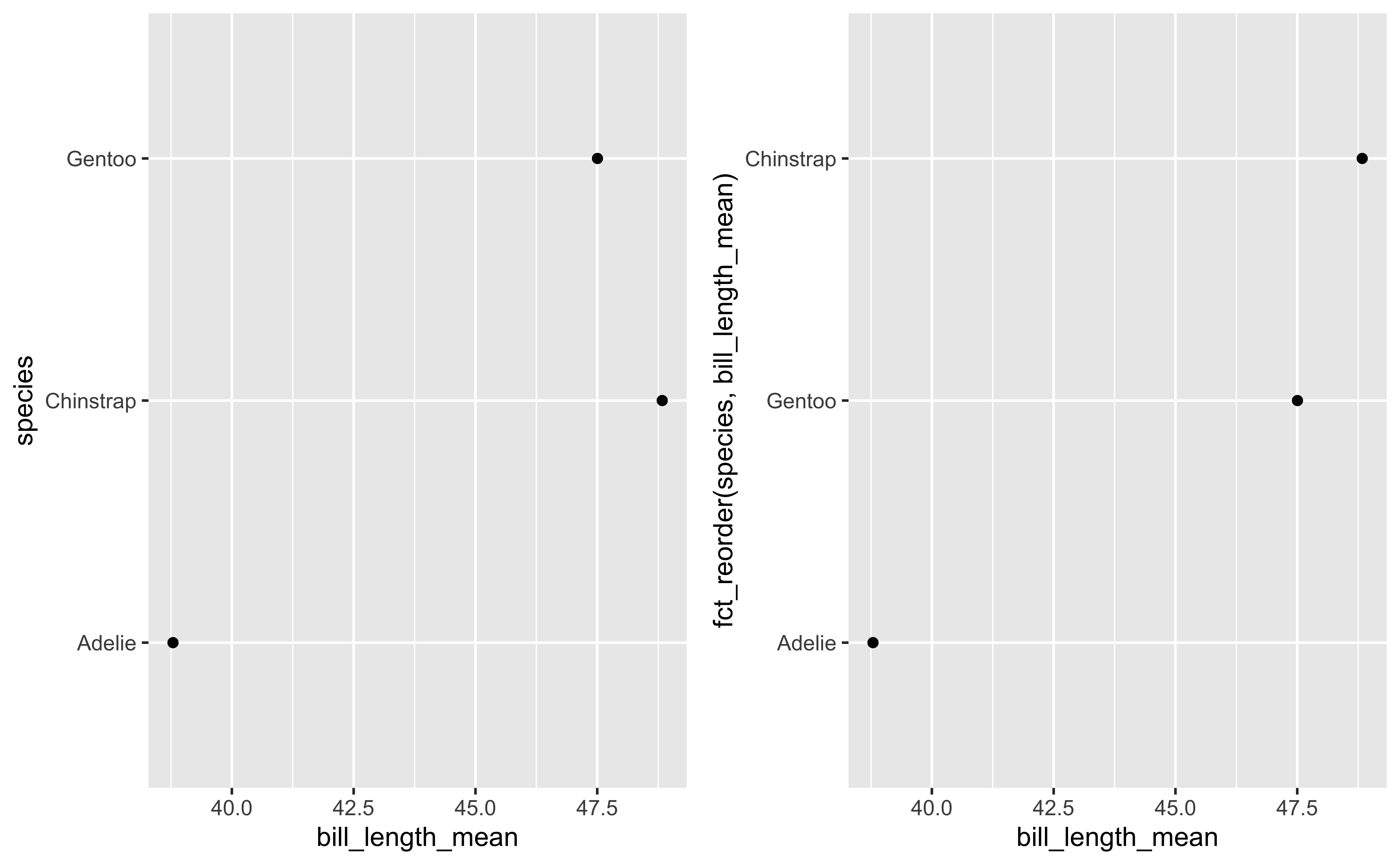
Examples for fct_reorder2
- Reorder the factor by the y values associated with the largest x values
- Easier to read: colours line up with the legend
fct_recode()
# A tibble: 3 × 2
species n
<fct> <int>
1 Adelie 152
2 Chinstrap 68
3 Gentoo 124penguins %>%
mutate(species = fct_recode(species,
"Adeliea" = "Adelie",
"ChinChin" = "Chinstrap",
"Gentooman" = "Gentoo")) %>%
count(species)# A tibble: 3 × 2
species n
<fct> <int>
1 Adeliea 152
2 ChinChin 68
3 Gentooman 124fct_collapse
Can use to collapse lots of levels
fct_lump
lubridate for dates and times
lubridate for dates and times
year(),month()mday()(day of the month),yday()(day of the year),wday()(day of the week)hour(),minute(),second()
Reference
- Allison Horst’s Posts
- Julie Scholler’s Slides
- R for Data Science
- Gina Reynolds’s Slides
- Sharla Gelfand’s Slides
- David’s Blog