# load packages
library(tidyverse)
library(tidymodels)
library(patchwork)
library(knitr)
library(kableExtra)
# set default theme and larger font size for ggplot2
ggplot2::theme_set(ggplot2::theme_minimal(base_size = 20))Model comparison
STA 210 - Summer 2022
Welcome
Announcements
- HW 2 posted today, due Sunday
- Lab 3 posted today, due Friday
Review on Exam 1 Part 1
Interpretation on coefficients
Topics
- ANOVA for Multiple Linear Regression and sum of squares
- Comparing models with \(R^2\) vs. \(R^2_{adj}\)
- Comparing models with AIC and BIC
- Occam’s razor and parsimony
- Overfitting and spending our data
Computational setup
Introduction
Data: Restaurant tips
Which variables help us predict the amount customers tip at a restaurant?
# A tibble: 169 × 4
Tip Party Meal Age
<dbl> <dbl> <chr> <chr>
1 2.99 1 Dinner Yadult
2 2 1 Dinner Yadult
3 5 1 Dinner SenCit
4 4 3 Dinner Middle
5 10.3 2 Dinner SenCit
6 4.85 2 Dinner Middle
7 5 4 Dinner Yadult
8 4 3 Dinner Middle
9 5 2 Dinner Middle
10 1.58 1 Dinner SenCit
# … with 159 more rowsVariables
Predictors:
Party: Number of people in the partyMeal: Time of day (Lunch, Dinner, Late Night)Age: Age category of person paying the bill (Yadult, Middle, SenCit)
Outcome: Tip: Amount of tip
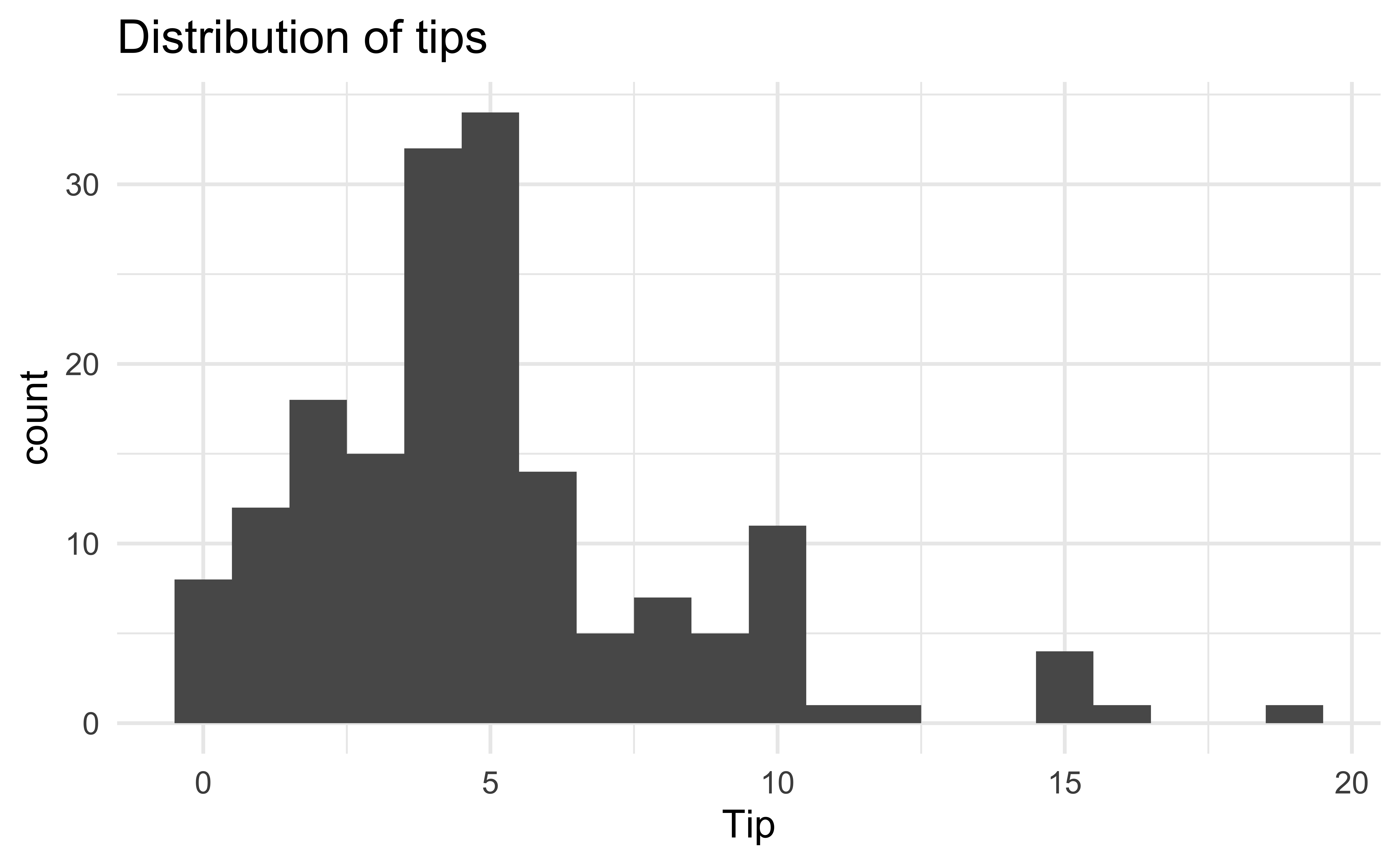
Outcome: Tip
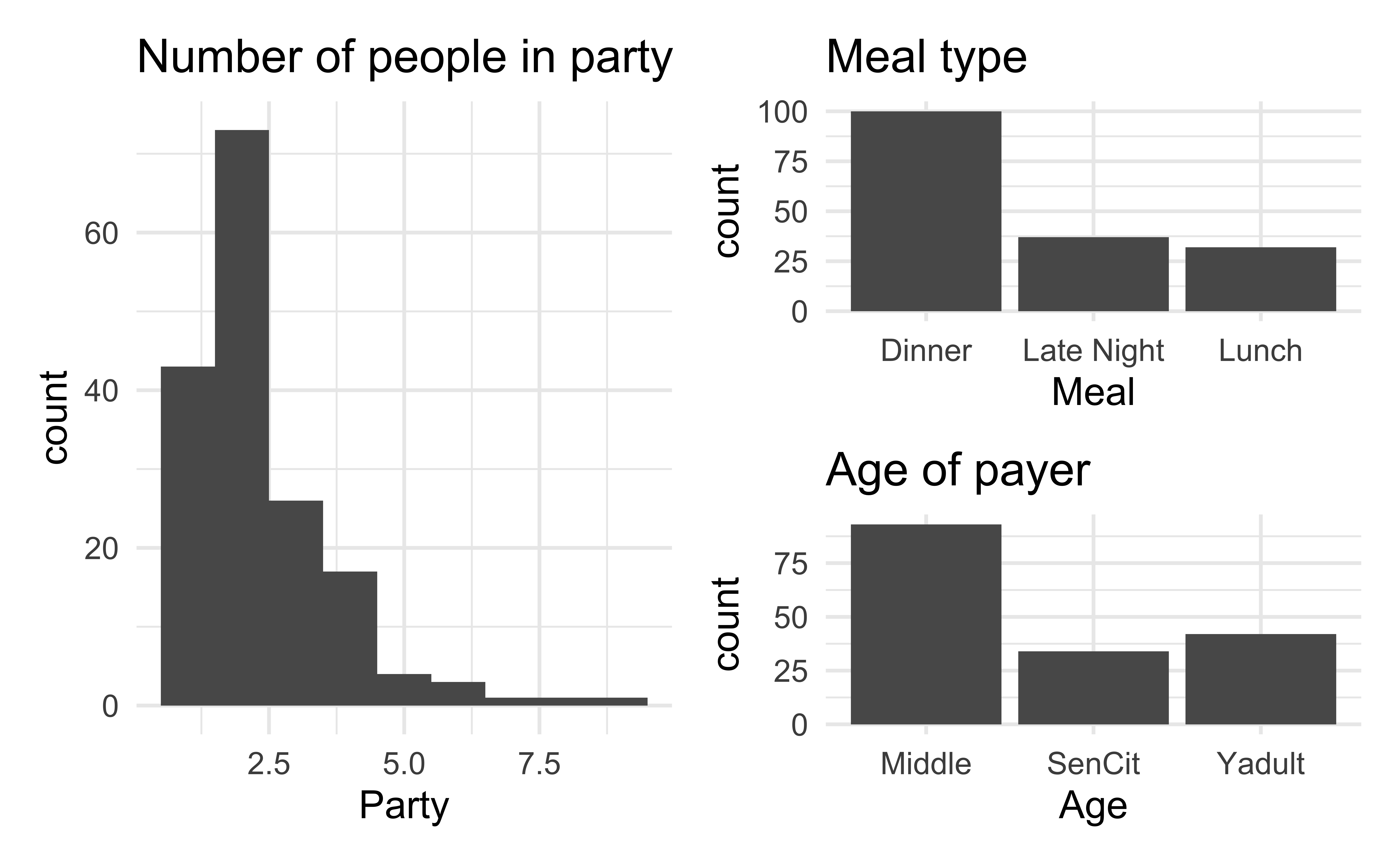
Predictors
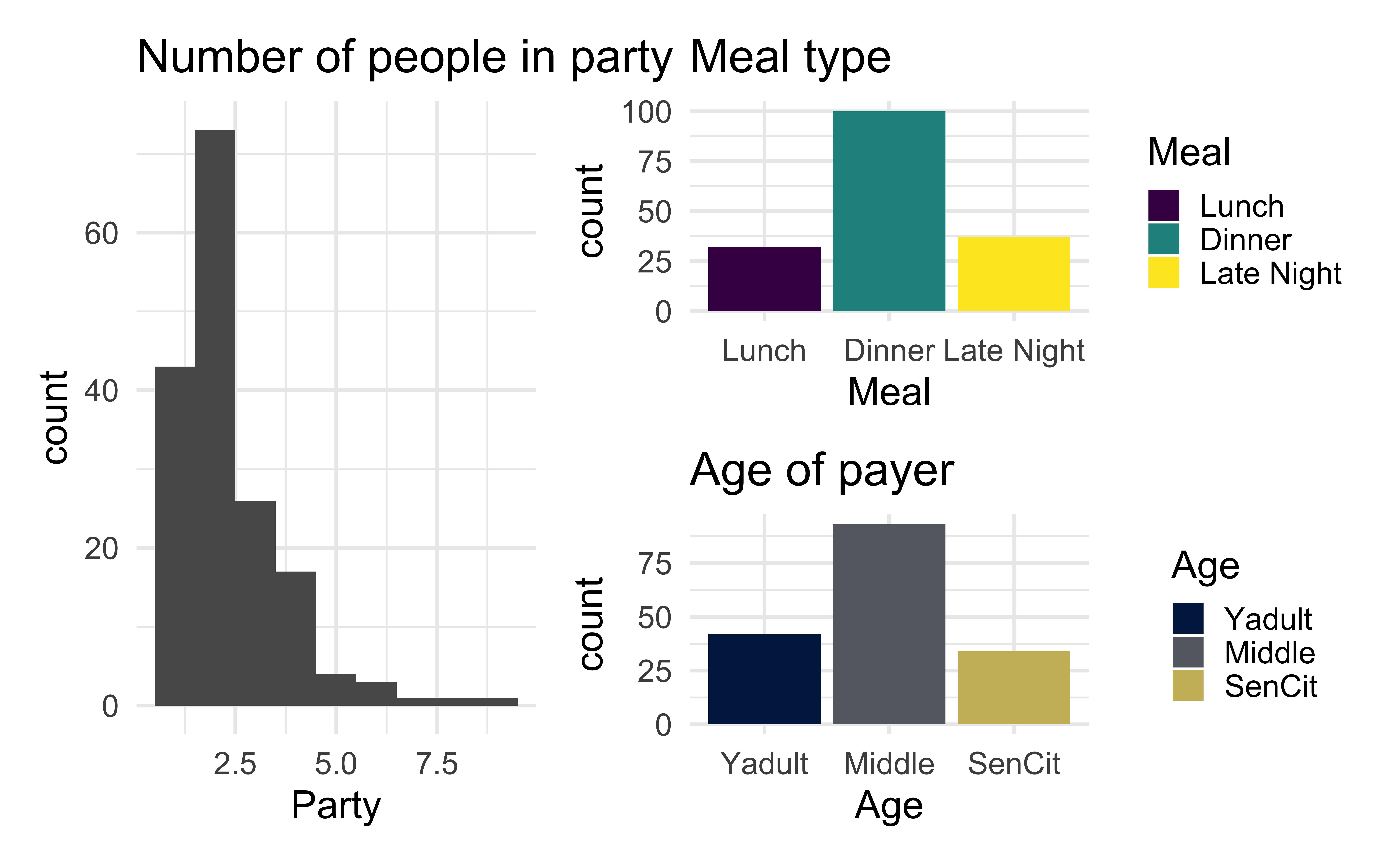
Relevel categorical predictors
tips <- tips %>%
mutate(
Meal = fct_relevel(Meal, "Lunch", "Dinner", "Late Night"),
Age = fct_relevel(Age, "Yadult", "Middle", "SenCit")
)Predictors, again
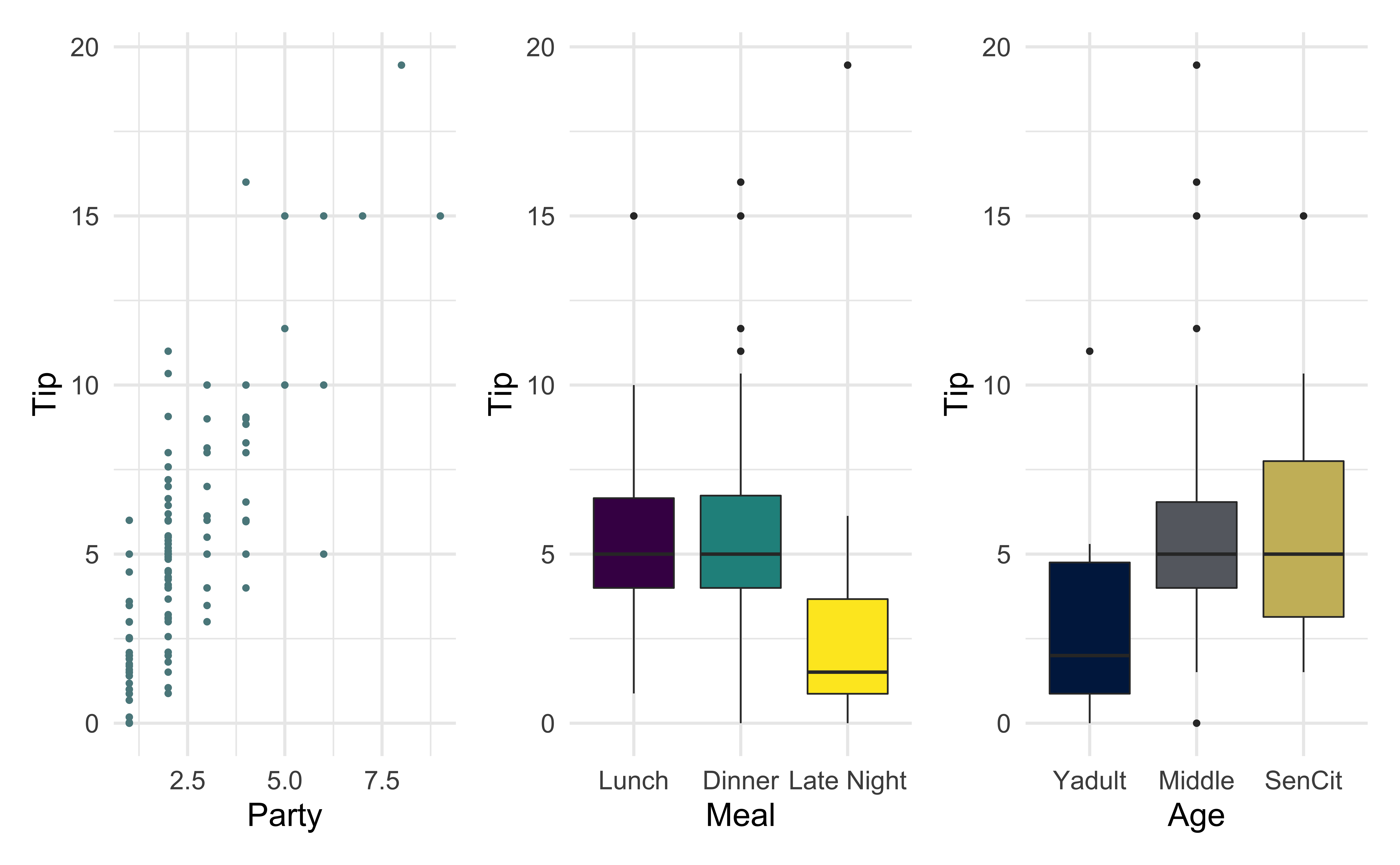
Outcome vs. predictors
Fit and summarize model
tip_fit <- linear_reg() %>%
set_engine("lm") %>%
fit(Tip ~ Party + Age, data = tips)
tidy(tip_fit, conf.int = TRUE) %>%
kable(digits = 3)| term | estimate | std.error | statistic | p.value | conf.low | conf.high |
|---|---|---|---|---|---|---|
| (Intercept) | -0.170 | 0.366 | -0.465 | 0.643 | -0.893 | 0.553 |
| Party | 1.837 | 0.124 | 14.758 | 0.000 | 1.591 | 2.083 |
| AgeMiddle | 1.009 | 0.408 | 2.475 | 0.014 | 0.204 | 1.813 |
| AgeSenCit | 1.388 | 0.485 | 2.862 | 0.005 | 0.430 | 2.345 |
Is this the best model to explain variation in tips?
Another model summary
| term | df | sumsq | meansq | statistic | p.value |
|---|---|---|---|---|---|
| Party | 1 | 1188.64 | 1188.64 | 285.71 | 0.00 |
| Age | 2 | 38.03 | 19.01 | 4.57 | 0.01 |
| Residuals | 165 | 686.44 | 4.16 | NA | NA |
Analysis of variance (ANOVA)
Analysis of variance (ANOVA)
- Main Idea: Decompose the total variation on the outcome into:
the variation that can be explained by the each of the variables in the model
the variation that can’t be explained by the model (left in the residuals)
- If the variation that can be explained by the variables in the model is greater than the variation in the residuals, this signals that the model might be “valuable” (at least one of the \(\beta\)s not equal to 0)
ANOVA output
anova(tip_fit$fit) %>%
tidy() %>%
kable(digits = 2)| term | df | sumsq | meansq | statistic | p.value |
|---|---|---|---|---|---|
| Party | 1 | 1188.64 | 1188.64 | 285.71 | 0.00 |
| Age | 2 | 38.03 | 19.01 | 4.57 | 0.01 |
| Residuals | 165 | 686.44 | 4.16 | NA | NA |
ANOVA output, with totals
| term | df | sumsq | meansq | statistic | p.value |
|---|---|---|---|---|---|
| Party | 1 | 1188.64 | 1188.64 | 285.71 | 0 |
| Age | 2 | 38.03 | 19.01 | 4.57 | 0.01 |
| Residuals | 165 | 686.44 | 4.16 | ||
| Total | 168 | 1913.11 |
Sum of squares
| term | df | sumsq |
|---|---|---|
| Party | 1 | 1188.64 |
| Age | 2 | 38.03 |
| Residuals | 165 | 686.44 |
| Total | 168 | 1913.11 |
- \(SS_{Total}\): Total sum of squares, variability of outcome, \(\sum_{i = 1}^n (y_i - \bar{y})^2\)
- \(SS_{Error}\): Residual sum of squares, variability of residuals, \(\sum_{i = 1}^n (y_i - \hat{y})^2\)
- \(SS_{Model} = SS_{Total} - SS_{Error}\): Variability explained by the model
R-squared, \(R^2\)
Recall: \(R^2\) is the proportion of the variation in the response variable explained by the regression model.
\[ R^2 = \frac{SS_{Model}}{SS_{Total}} = 1 - \frac{SS_{Error}}{SS_{Total}} = 1 - \frac{686.44}{1913.11} = 0.641 \]
glance(tip_fit)# A tibble: 1 × 12
r.squared adj.r.squared sigma statistic p.value df logLik AIC BIC
<dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
1 0.641 0.635 2.04 98.3 1.56e-36 3 -358. 726. 742.
# … with 3 more variables: deviance <dbl>, df.residual <int>, nobs <int>Model comparison
R-squared, \(R^2\)
- \(R^2\) will always increase as we add more variables to the model + If we add enough variables, we can always achieve \(R^2=100\%\)
- If we only use \(R^2\) to choose a best fit model, we will be prone to choose the model with the most predictor variables
Adjusted \(R^2\)
- Adjusted \(R^2\): measure that includes a penalty for unnecessary predictor variables
- Similar to \(R^2\), it is a measure of the amount of variation in the response that is explained by the regression model
- Differs from \(R^2\) by using the mean squares rather than sums of squares and therefore adjusting for the number of predictor variables
\(R^2\) and Adjusted \(R^2\)
\[R^2 = \frac{SS_{Model}}{SS_{Total}} = 1 - \frac{SS_{Error}}{SS_{Total}}\]
\[R^2_{adj} = 1 - \frac{SS_{Error}/(n-p-1)}{SS_{Total}/(n-1)}\]
Using \(R^2\) and Adjusted \(R^2\)
- Adjusted \(R^2\) can be used as a quick assessment to compare the fit of multiple models; however, it should not be the only assessment!
- Use \(R^2\) when describing the relationship between the response and predictor variables
Comparing models with \(R^2_{adj}\)
tip_fit_1 <- linear_reg() %>%
set_engine("lm") %>%
fit(Tip ~ Party +
Age +
Meal,
data = tips)
glance(tip_fit_1) %>%
select(r.squared, adj.r.squared)# A tibble: 1 × 2
r.squared adj.r.squared
<dbl> <dbl>
1 0.674 0.664tip_fit_2 <- linear_reg() %>%
set_engine("lm") %>%
fit(Tip ~ Party +
Age +
Meal +
Day,
data = tips)
glance(tip_fit_2) %>%
select(r.squared, adj.r.squared)# A tibble: 1 × 2
r.squared adj.r.squared
<dbl> <dbl>
1 0.683 0.662AIC & BIC
Estimators of prediction error and relative quality of models:
Akaike’s Information Criterion (AIC): \[AIC = n\log(SS_\text{Error}) - n \log(n) + 2(p+1)\]
Schwarz’s Bayesian Information Criterion (BIC): \[BIC = n\log(SS_\text{Error}) - n\log(n) + log(n)\times(p+1)\]
AIC & BIC
\[ \begin{aligned} & AIC = \color{blue}{n\log(SS_\text{Error})} - n \log(n) + 2(p+1) \\ & BIC = \color{blue}{n\log(SS_\text{Error})} - n\log(n) + \log(n)\times(p+1) \end{aligned} \]
First Term: Decreases as p increases
AIC & BIC
\[ \begin{aligned} & AIC = n\log(SS_\text{Error}) - \color{blue}{n \log(n)} + 2(p+1) \\ & BIC = n\log(SS_\text{Error}) - \color{blue}{n\log(n)} + \log(n)\times(p+1) \end{aligned} \]
Second Term: Fixed for a given sample size n
AIC & BIC
\[ \begin{aligned} & AIC = n\log(SS_\text{Error}) - n\log(n) + \color{blue}{2(p+1)} \\ & BIC = n\log(SS_\text{Error}) - n\log(n) + \color{blue}{\log(n)\times(p+1)} \end{aligned} \]
Third Term: Increases as p increases
Using AIC & BIC
\[ \begin{aligned} & AIC = n\log(SS_{Error}) - n \log(n) + \color{red}{2(p+1)} \\ & BIC = n\log(SS_{Error}) - n\log(n) + \color{red}{\log(n)\times(p+1)} \end{aligned} \]
Choose model with the smaller value of AIC or BIC
If \(n \geq 8\), the penalty for BIC is larger than that of AIC, so BIC tends to favor more parsimonious models (i.e. models with fewer terms)
Comparing models with AIC and BIC
tip_fit_1 <- linear_reg() %>%
set_engine("lm") %>%
fit(Tip ~ Party +
Age +
Meal,
data = tips)
glance(tip_fit_1) %>%
select(AIC, BIC)# A tibble: 1 × 2
AIC BIC
<dbl> <dbl>
1 714. 736.tip_fit_2 <- linear_reg() %>%
set_engine("lm") %>%
fit(Tip ~ Party +
Age +
Meal +
Day,
data = tips)
glance(tip_fit_2) %>%
select(AIC, BIC)# A tibble: 1 × 2
AIC BIC
<dbl> <dbl>
1 720. 757.Commonalities between criteria
- \(R^2_{adj}\), AIC, and BIC all apply a penalty for more predictors
- The penalty for added model complexity attempts to strike a balance between underfitting (too few predictors in the model) and overfitting (too many predictors in the model)
- Goal: Parsimony
Parsimony and Occam’s razor
The principle of parsimony is attributed to William of Occam (early 14th-century English nominalist philosopher), who insisted that, given a set of equally good explanations for a given phenomenon, the correct explanation is the simplest explanation1
Called Occam’s razor because he “shaved” his explanations down to the bare minimum
Parsimony in modeling:
- models should have as few parameters as possible
- linear models should be preferred to non-linear models
- experiments relying on few assumptions should be preferred to those relying on many
- models should be pared down until they are minimal adequate
- simple explanations should be preferred to complex explanations
In pursuit of Occam’s razor
Occam’s razor states that among competing hypotheses that predict equally well, the one with the fewest assumptions should be selected
Model selection follows this principle
We only want to add another variable to the model if the addition of that variable brings something valuable in terms of predictive power to the model
In other words, we prefer the simplest best model, i.e. parsimonious model
Alternate views
Sometimes a simple model will outperform a more complex model . . . Nevertheless, I believe that deliberately limiting the complexity of the model is not fruitful when the problem is evidently complex. Instead, if a simple model is found that outperforms some particular complex model, the appropriate response is to define a different complex model that captures whatever aspect of the problem led to the simple model performing well.
Radford Neal - Bayesian Learning for Neural Networks2
Other concerns with our approach
- All criteria we considered for model comparison require making predictions for our data and then uses the prediction error (\(SS_{Error}\)) somewhere in the formula
- But we’re making prediction for the data we used to build the model (estimate the coefficients), which can lead to overfitting
- Instead we should
split our data into testing and training sets
“train” the model on the training data and pick a few models we’re genuinely considering as potentially good models
test those models on the testing set
Recap
ANOVA for Multiple Linear Regression and sum of squares
Comparing models with \(R^2\) vs. \(R^2_{adj}\)
Comparing models with AIC and BIC
Occam’s razor and parsimony
Overfitting and spending our data